scRNA seq-guided data
for a successful GEX single-cell RNA sequencing (scRNA-seq) or gene expression analysis, for gene expression (GEX) studies and other analyses like proteomics or metabolomics
Instrument Software SeekOne SeekSoul
®
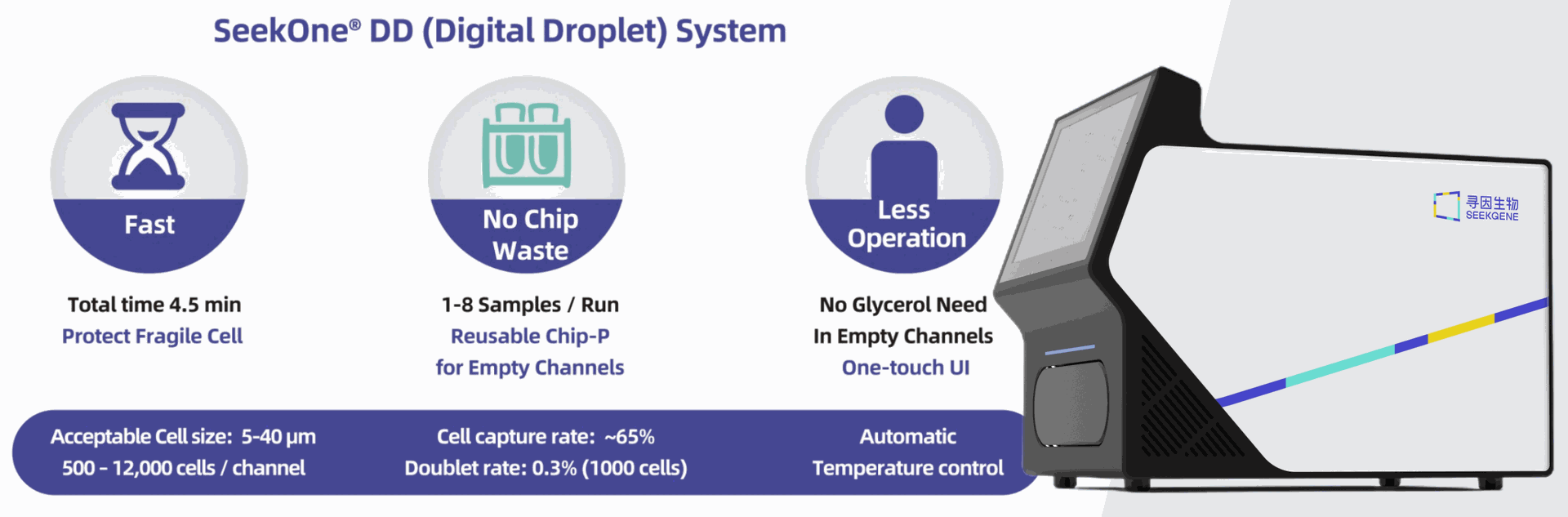
Tools Digital Droplet System SeekOne DD
®
Microfluidic Control SystemSeekMate Tinitan
TM
Fluoresence Cell Counter SeekOne
®
DD Single Cell 3’Transcriptome Kit SeekOne
®
DD Single Cell Immune Profiling Kit SeekOne
®
DD Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit
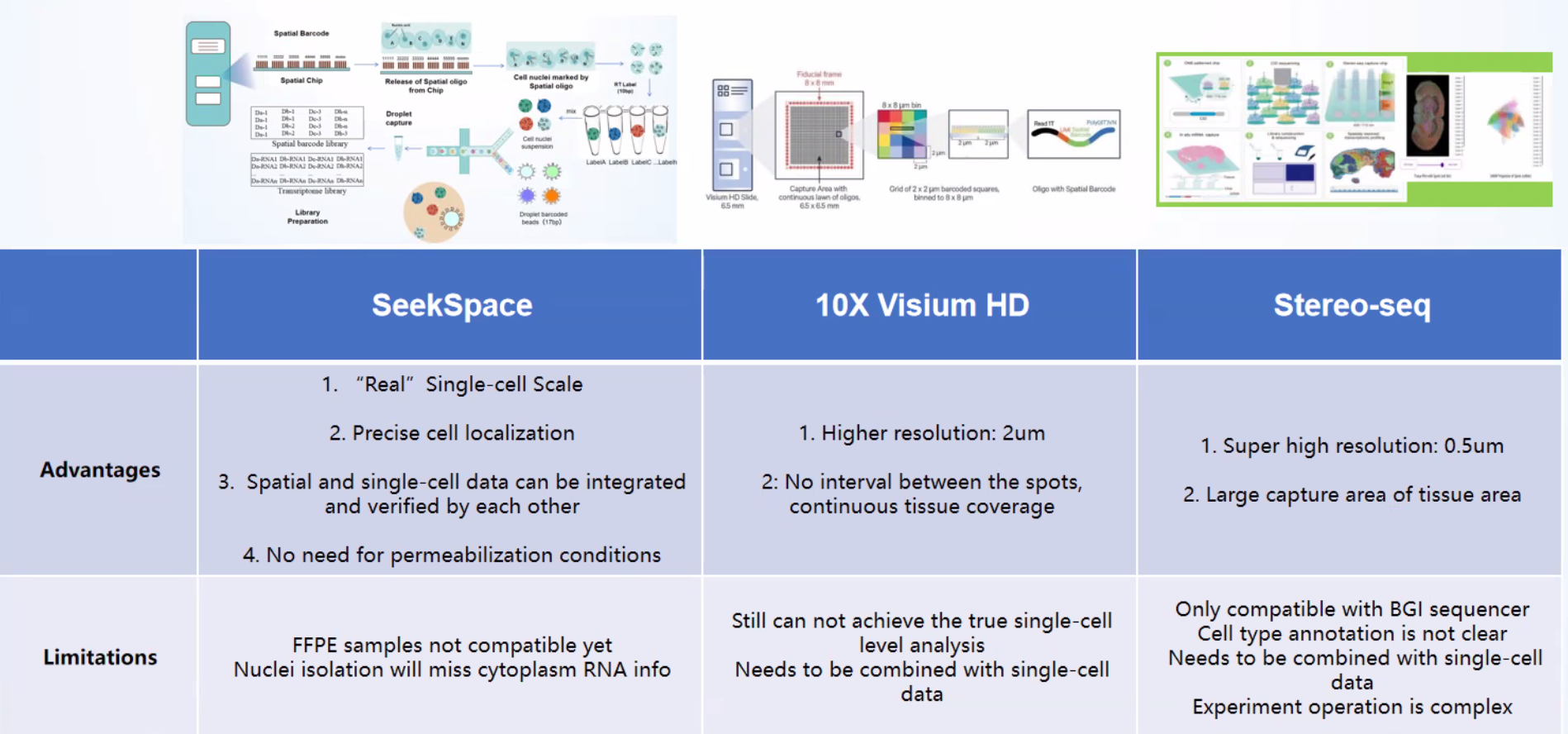
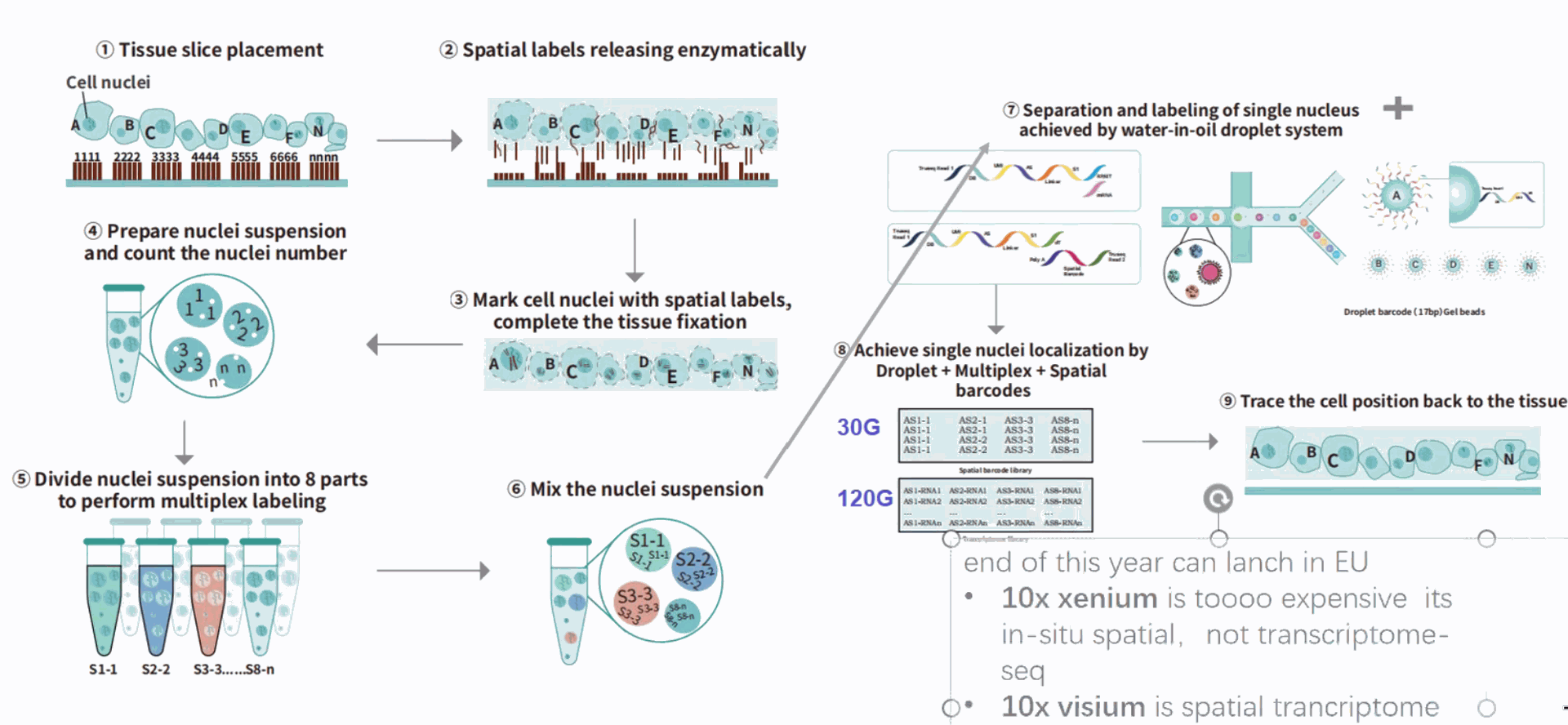

Approach
The products and services offered are
|
SeekOne® Digital Droplet System |
M001A |
1 unit |
unit |
|
SeekSpace® High-Frequency Ultrasonic Device |
M005A |
1 unit |
unit |
|
SeekMate Tinitan® Fluorescence Cell Counter |
M002C |
1 unit |
unit |
Reagent kit
|
Product Name |
Model |
Specification |
Unit |
SeekOne® DD Single Cell 3' Transcriptome-seq Kit |
K00202-08 |
8 tests |
set |
SeekOne® DD Single Cell 5' Transcriptome-seq Kit |
K00501-08 |
8 tests |
set |
SeekOne® DD Single Cell TCR Enrichment Kit (Human) |
K00601-08 |
8 tests |
set |
SeekOne® DD Single Cell BCR Enrichment Kit (Human) |
K00701-08 |
8 tests |
set |
SeekOne® DD Single Cell TCR Enrichment Kit (Mouse) |
K01101-08 |
8 tests |
set |
SeekOne® DD Single Cell BCR Enrichment Kit (Mouse) |
K01201-08 |
8 tests |
set |
SeekOne® DD Single Cell Full-length RNA Sequence Transcriptome-seq Kit |
K00801-08 |
8 tests |
set |
SeekOne® DD FFPE Single Cell Transcriptome-seq Kit |
K02101-08 |
8 tests |
set |
SeekMate FFPE Dissociation Kit |
K02301-08 |
8 tests |
set |
SeekSpace® Single Cell Spatial Transcriptome-seq Kit |
K02501-08 |
8 tests |
set |
SeekSpace® Single Cell Spatial Transcriptome-seq Kit |
K02501-02 |
2 tests |
set |
SeekSpace® Chip Holder |
SP00054 |
1 unit |
unit |
SeekMate® Tissue Dissociation Kit A Pro |
K01801-08 |
8 tests |
set |
SeekMate® Tissue Dissociation Kit A Pro |
K01801-30 |
30 tests |
set |
SeekMate Cell Counter Chip |
SP00022 |
50 pieces |
box |
SeekMate AO/PI Staining Kit |
K01701 |
5 ml |
bottle |
. This content is the foundation of our products and services and is managed in SeekBase, the knowledge core of SeekQuence.
The extensive body of knowledge includes reagents and tools used in the scientific world, what research applications they were used for, by what researchers, purchased from which suppliers, and published in what peer-reviewed publications. Applying computational and analytical methods, SeekQuence established relationships between these different entities.

Products and Services
Product search portal
Expedite experimental design
For the researcher in academia and industry
One year trial on one unit of SeekOne® Digital Droplet System, REF: M001A 9000 Euro
SeekOne Single Cell 3’ Transcriptomeseq Kit, 8 tests per kit REF: K00202-08 9000 Euro
SeekProducts is a product search portal, designed with the researcher in mind that needs to quickly identify the right product for the next experiment.
Design the next experiment with a trusted and validated product. Knowing that a product has been successfully used for a specific scientific application by respected scientists, as demonstrated by a peer-reviewed publication, is of great importance to any researcher.
- SURFSeq 5000* Sequencing Platform
- FASTASeq 300* Sequencing Platform
- GenoLab M* Sequencing Platform
- MrLH-96
Automated Sample Preparation System
Search across a vast amount of product suppliers - supplier-agnostic
Find products based on research area of interest
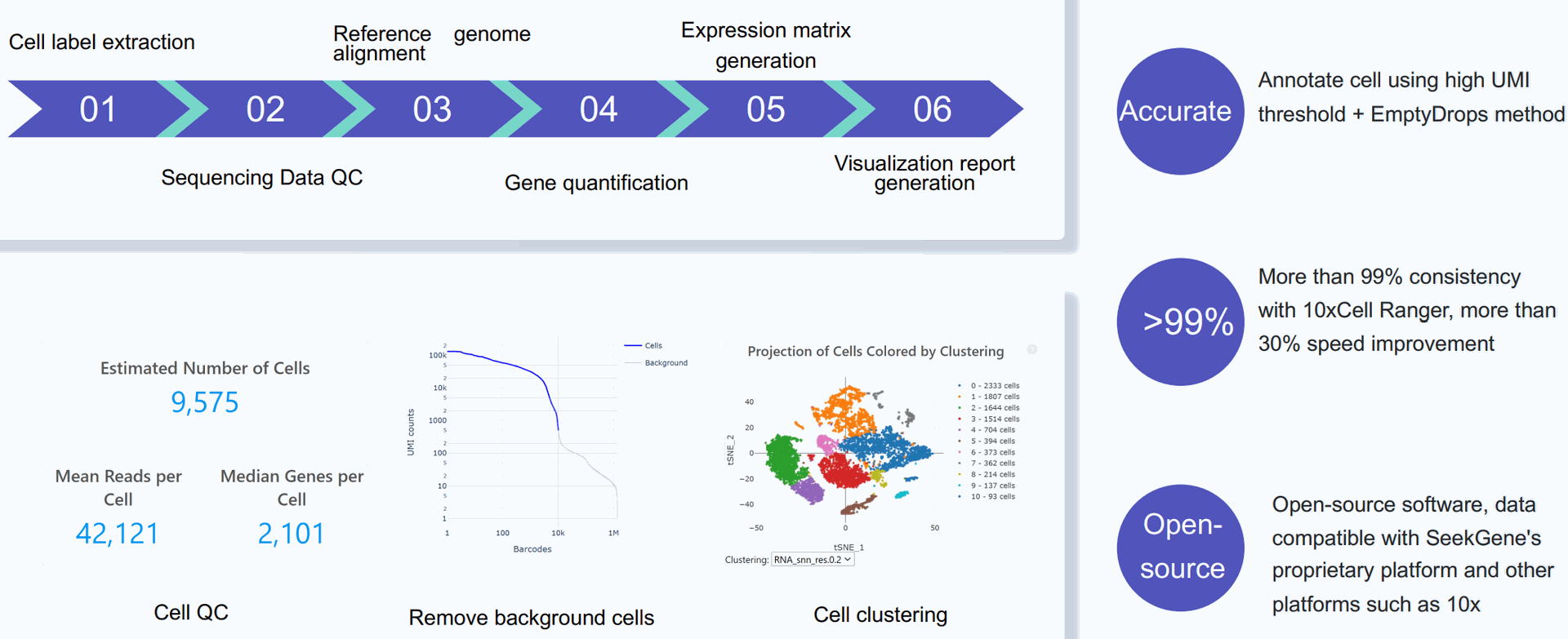
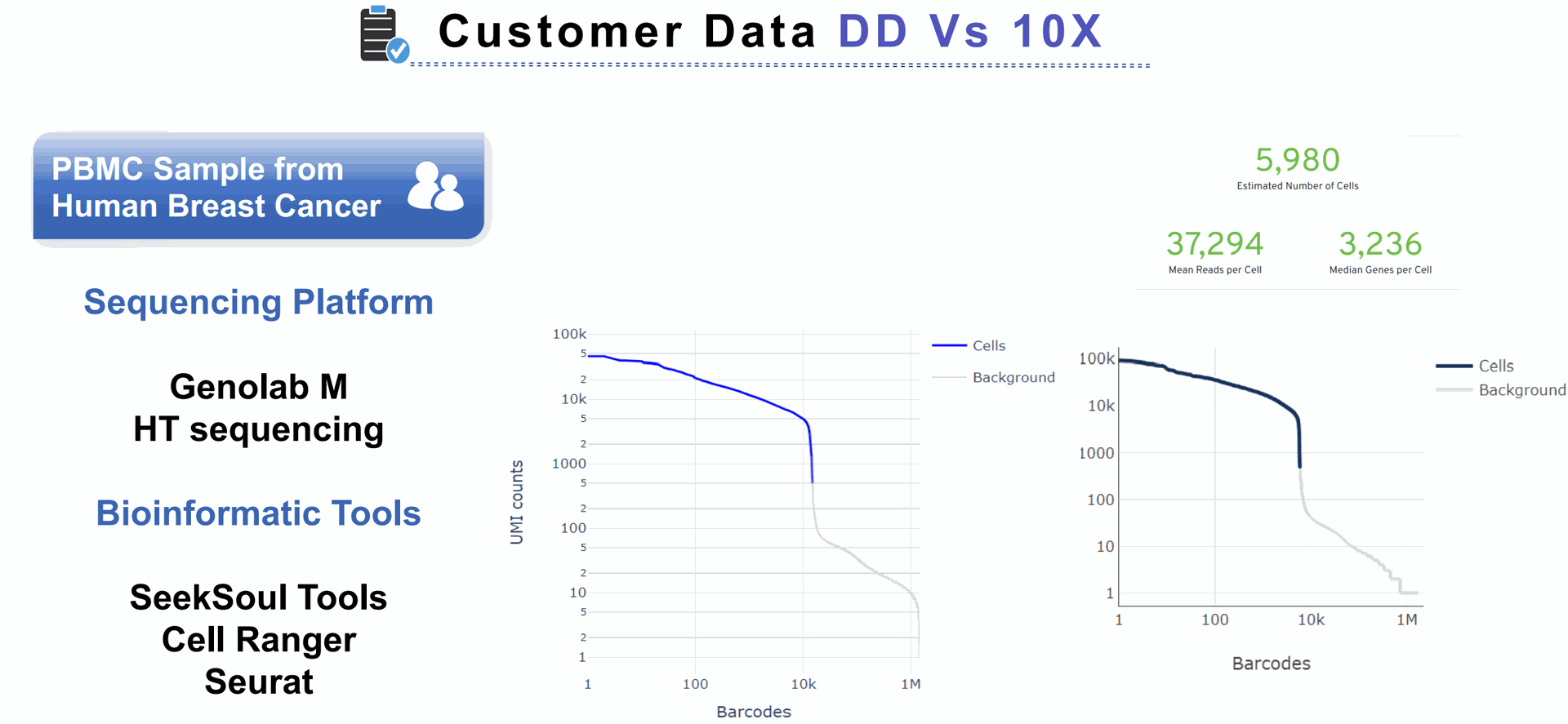
UMI counts represent the absolute number of observed transcripts (per gene, cell or sample). As absolute counts, they are not normalized for sequencing depth, technical variation or RNA content per cell.
Filter down to relevant products via a comprehensive set of facets
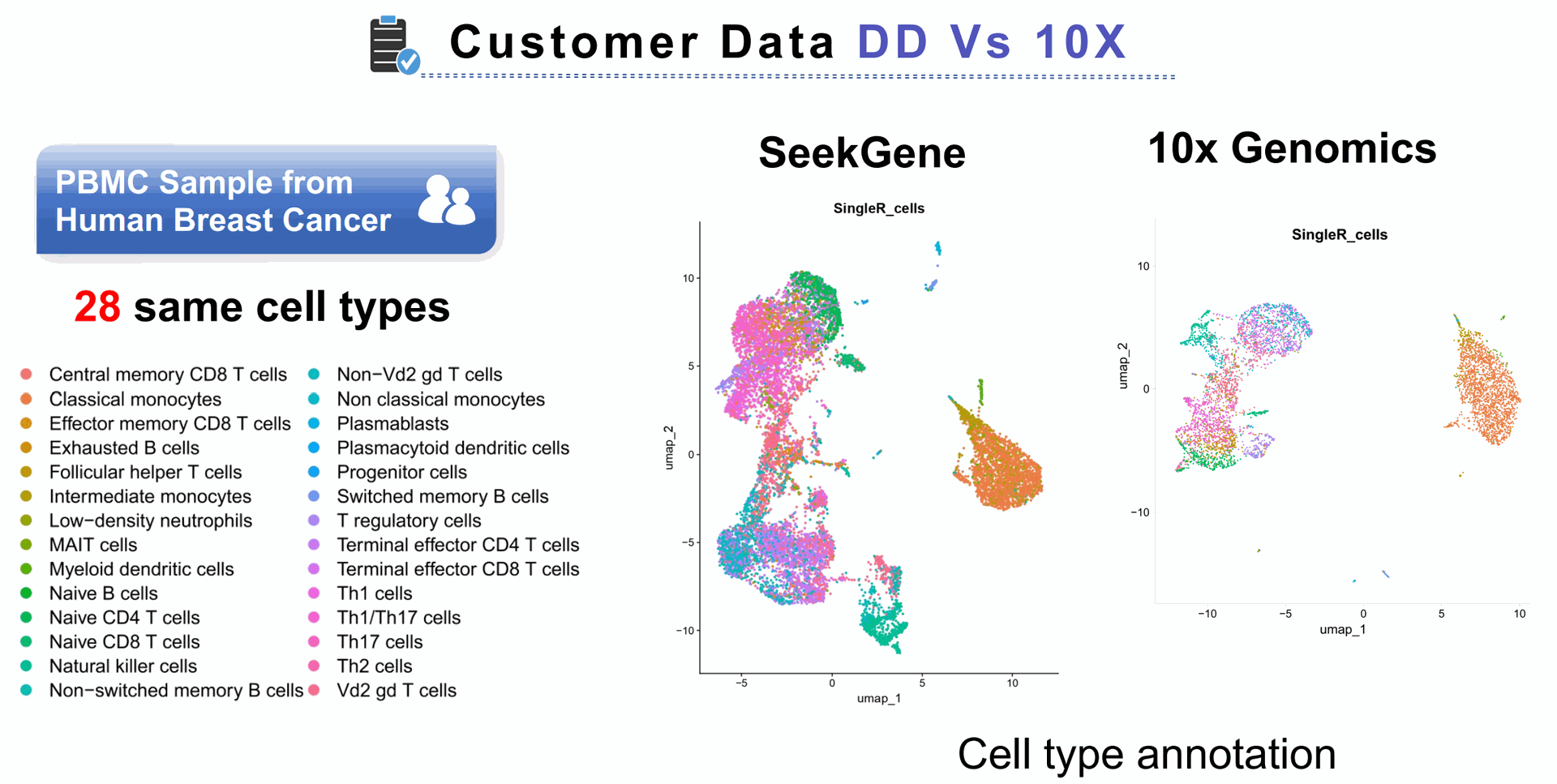
Quickly identify any publication and researcher that has successfully used the scRNA Digital Droplet seq strategy of interest with longest reads like Seekgene
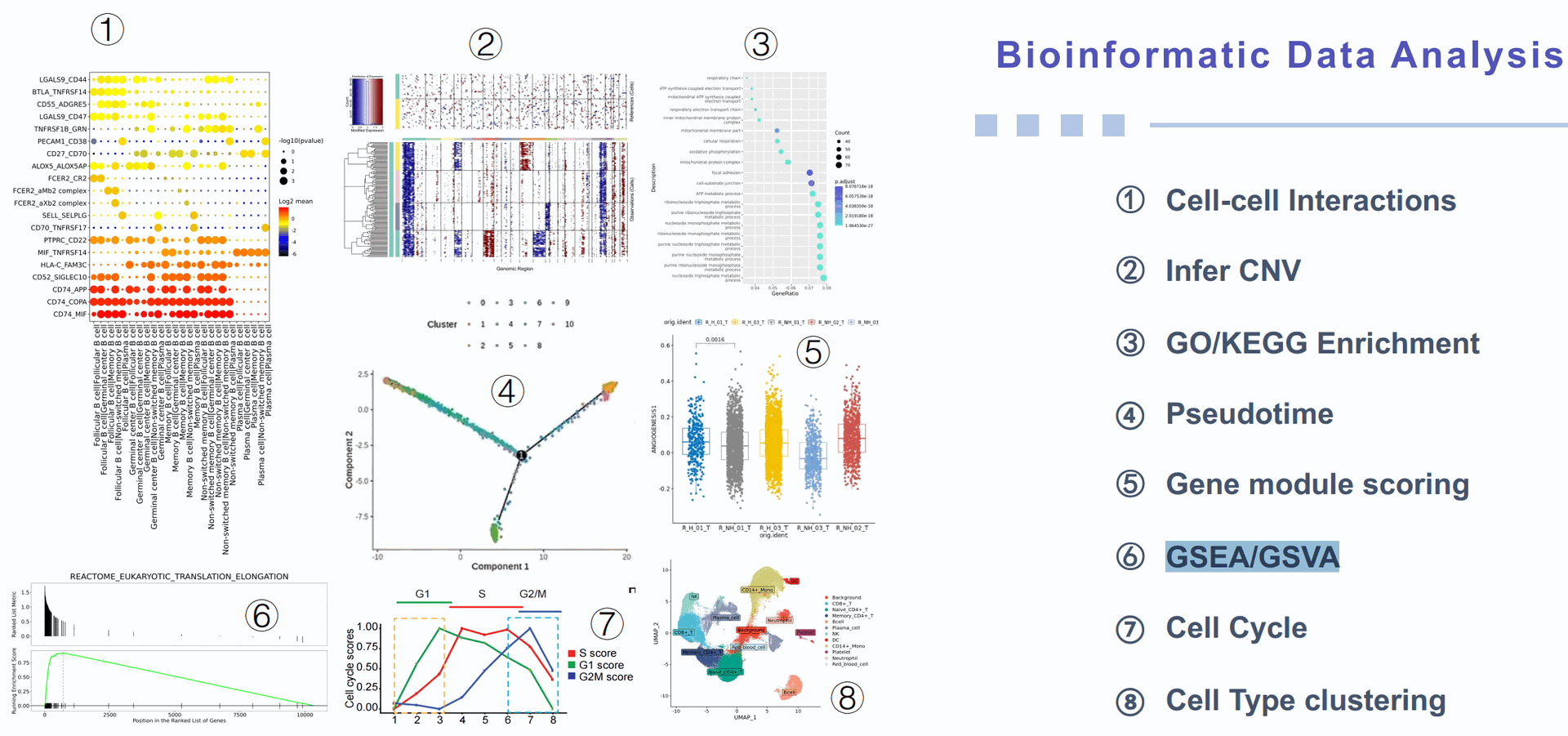
Find relevant GSEA that employs differentially expressed genes between sample groups for functional enrichment analysis, whereas GSVA estimates the variations in gene set enrichment for each sample based on the expression profiles to elucidate functional enrichment differences between sample groups
Cloud Analysis with SeekSoul a web-based application that is stronger than Cell Ranger and Space Ranger from 10x Genomics' cloud.
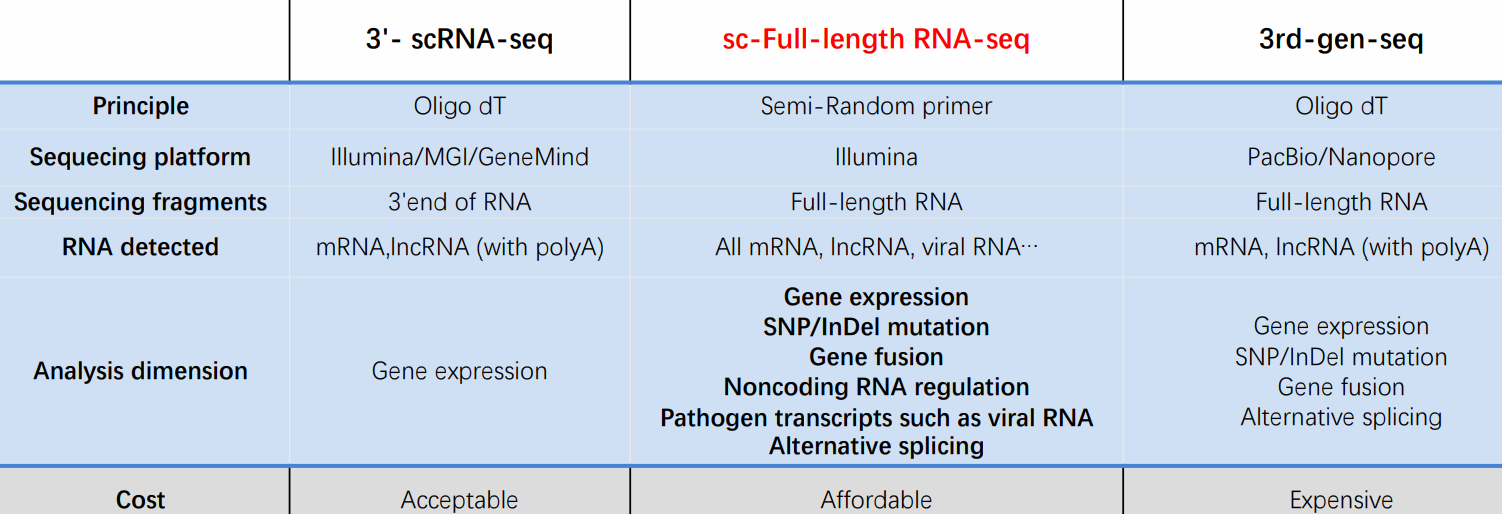
SeekOne DD scFAST-seq is better than the Nanopore 3th generation
- Digital Droplet scFAST seq can analyse mRNA, lncRNA, viral RNA, mRNA, lncRNA (with polyA) Analysis dimension Gene expression Gene expression SNP/InDel mutation Gene fusion Non coding RNA regulation Pathogen transcripts such as viral RNA Alternative splicing.
- 3rd-gen-seq with PacBio/Nanopore is more expensive and has more data limitations than Digital Droplet scFAST seq analyses mRNA
- BOTH CAN ANALYSE Long non-coding RNA (lncRNA) are RNA transcripts over 200 nucleotides in length that do not encode proteins. This is a broad definition, encompassing around 270 000 unique RNA in humans, although only 15000 have so far been functionally annotated
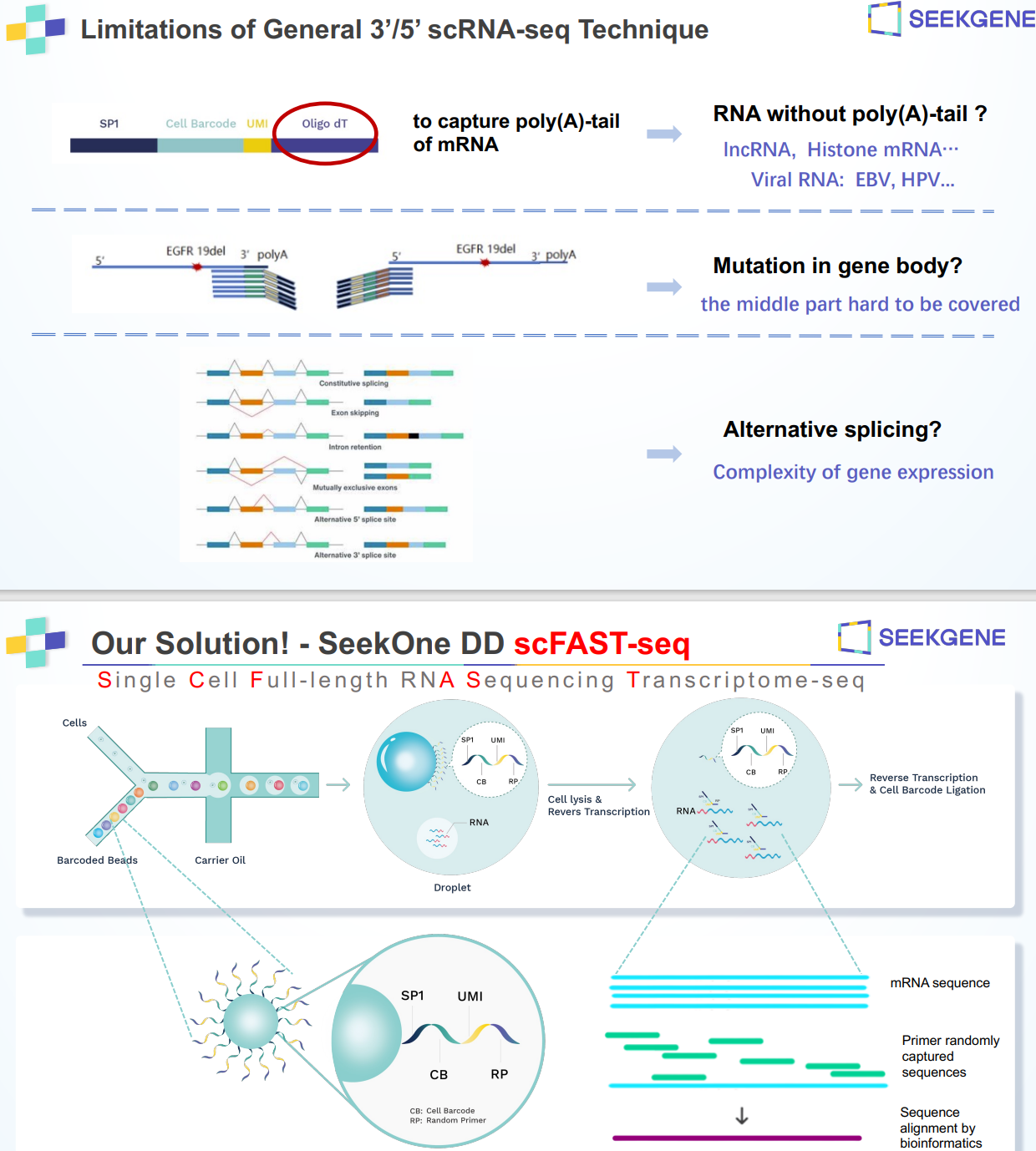
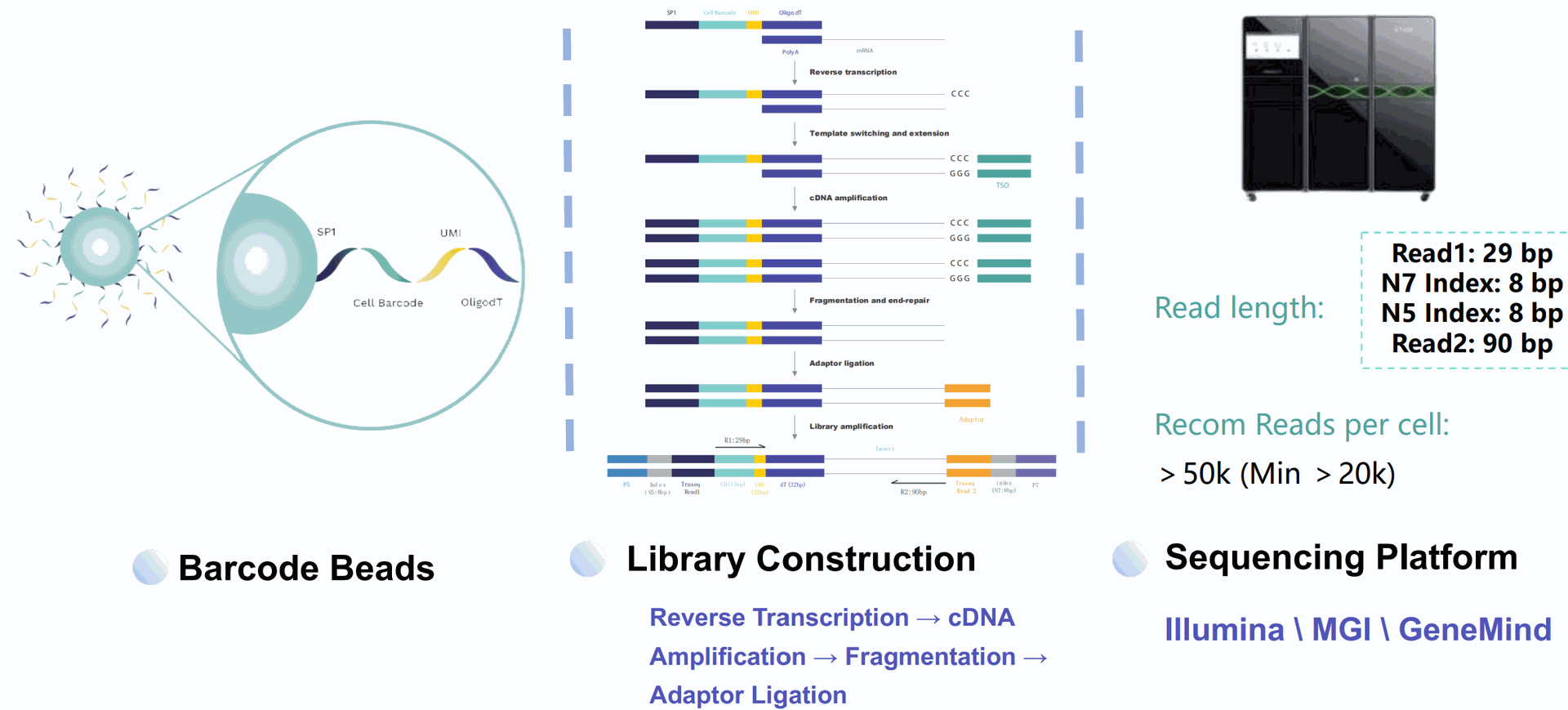
SeekOne Digital Droplet (SeekOne DD) Higg throughput Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit
High-throughput whole transcriptome profiling. The scFAST-seq method makes use of innovative techniques
including semi-random primers, efficient reverse transcription, template swapping, and effective rRNA removal to build full-length RNA libraries of up to 12,000 cells. Compared to conventional 3' scRNA-seq, scFAST- seq has distinct advantages in detecting non- polyadenylated transcripts, transcript coverage length, and identification of more splice junctions. With target region enrichment, scFAST-seq can simultaneously detect somatic mutations and cell states in individual tumour cells, providing valuable information for precision medicine. The kit is designed to be used in conjunction with our SeekOne Digital Droplet System (SeekOne DD)
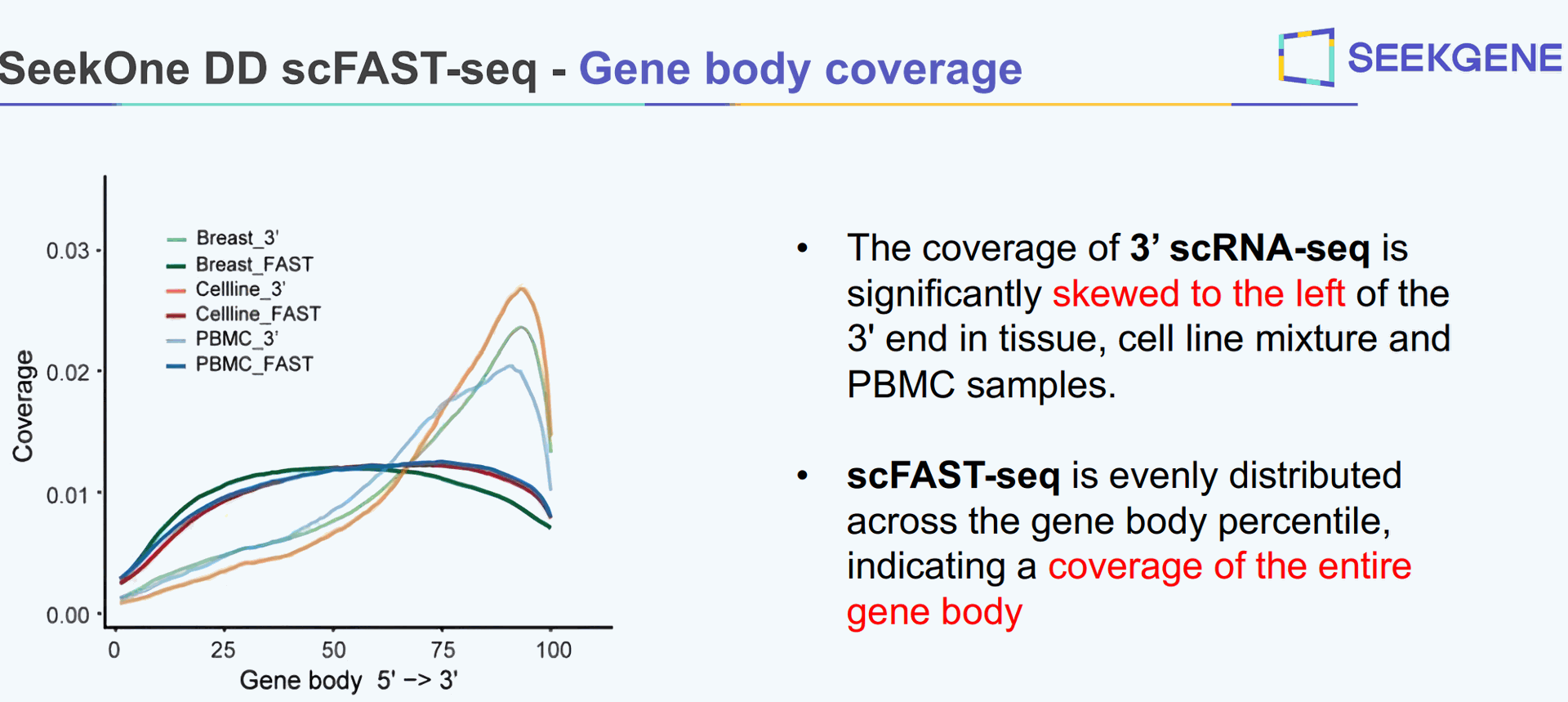
We compared SeekOne DD scFAST seq with the 3' 10x single cell data from a 3' expression kit. The poly(dT)VN captures the end of the transcript and one can sequence 150 bp. While visualizing BAM file in IGV. I filtered the BAM file to only include reads used for UMI counting.
You will only be sequencing 150 bp and only expect to see reads aligning to maybe the last two exons of a transcript?
That is why Seekgene offers to complete the entire process from single-cell nucleic acid labelling to transcriptome library construction. When equipped with "SeekSoul Tools", our single-cell data analysis software, we provide a one-stop-shop single-cell transcriptome solution.
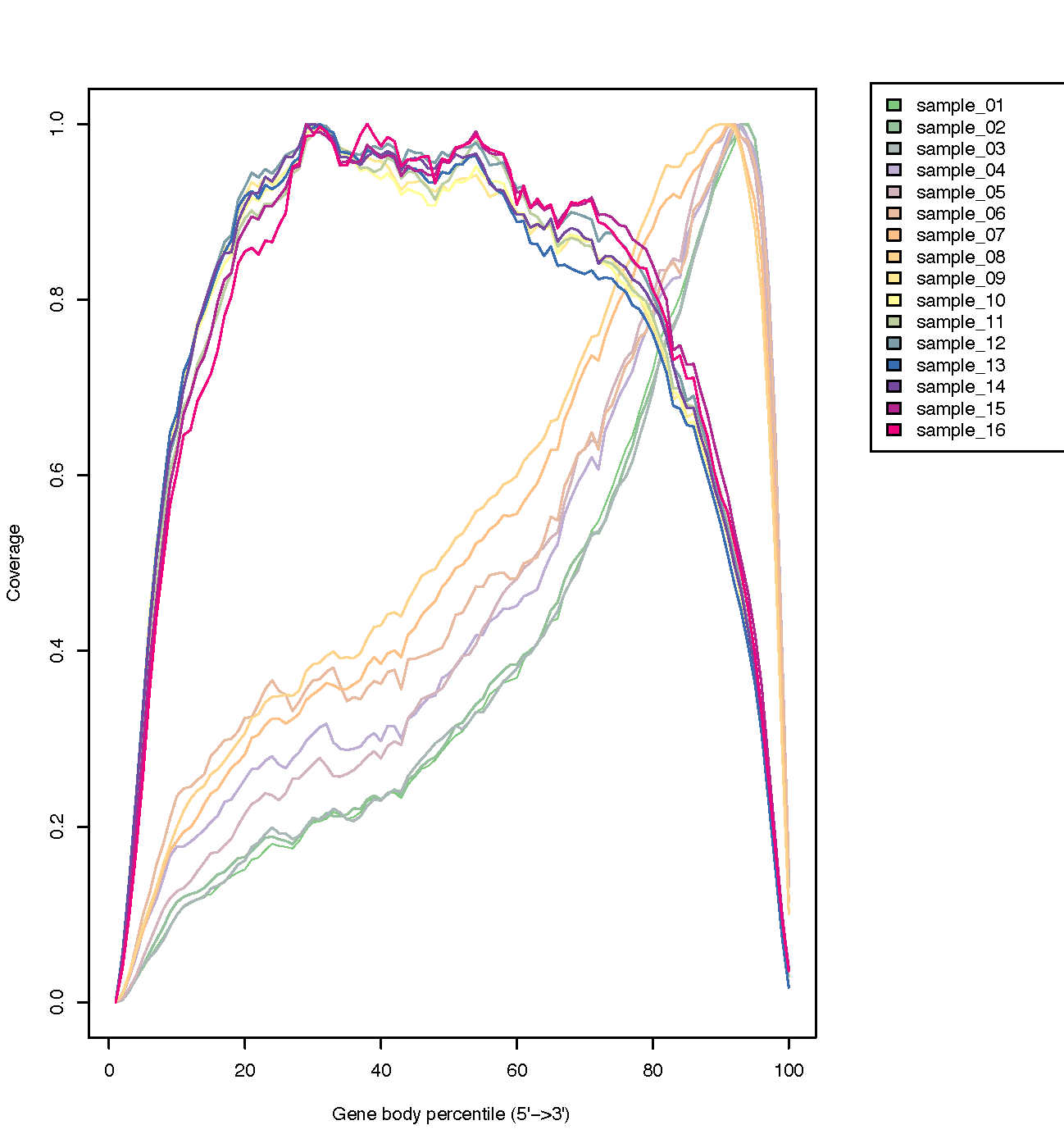
Functions of the RSeQC tool are called (incl. the gene coverage plot)
Having said this, in the past I came across an R package called genecovr (link) that based on the available info might do what you are looking for ("The genecovr package provides functionality for evaluating gene body coverage and other metrics in genome assemblies. The central measurement unit is a gene or transcript model"). Also see the vignette.
Also, I know that the R package accompanying the QoRTs tool is used to make visualizations of QC metrics, including gene coverage plots. Again, I don't have any hands-on experience with it, so I cannot comment on its compatibility with your output files...
<<forgot to add>> When browsing Bioconductor packages I also noticed the package CoverageView. Although primarily designed for the visualization of ChIp-seq-like experiments, "additionally, CoverageView can also be used in RNA-seq experiments in order to compare how the coverage is distributed along the transcriptome."
Azenta
Bio Molecular Systems
Biolidics
Bioptic Inc
Bioscreen
Byonoy
Catch Gene
CEM
Denovix
- DS-11: Spectrophotometer, cuvette and fluorimeter
- DS-8X: Eight-channel spectrophotometer
- DS-7: UV-Vis Spectrophotometer
- CellDrop™: Automated Cell Counter
- QFX Fluorometer
- Fluorescence quantification kits
- Cell counting tests
Gesim
Iota Sciences
Levitas Bio
Nano Analytics
- CellZscope 2: TEER measurement
- CellZscope +: TEER measurement
- CellZscope E: TEER measurement
- Cellzscope 3: TEER measurement
Nicoya
PAA – Peak Analysis & Automation
Porvair Sciences
- UltraVap Evaporator
- Ultraseal™ Heat Sealer
- Labeler and Plate Handler
- Deepwell Microplate for KingFisher™ Systems
Proteome Software
RBC Bioscience
- Magcore Plus II: CE-IVD DNA Extractor
- MagCore® EDA: PCR Extractor and Preparer
- MagCore Extraction Kits
S2 Genomics
SeekGene
Yokogawa
Zybio
Find antibodies for different model organism - beyond human antibodies
Initial focus is antibody products, but other product categories (e.g. NGS products) are coming soon.