SeekOne DD Single Cell Immune Profiling
Platform
Single-cell immune repertoire sequencing, also known
as single-cell V(D)J sequencing, involves targeting
single T lymphocytes or B lymphocytes, constructing
an immune repertoire RNA library via RNA enrichment
of the TCR (T-cell receptor)/BCR (B-cell receptor)
region, and analyzing the diverse sequences of
TCR/BCR, to obtain the immune features of the body.
SeekOne® DD single-cell immune profiling (5'
transcriptome & immune repertoire sequencing),
proposed by Beijing SeekGene BioSciences Co., Ltd.,
achieves single-cell partitioning and labelling by the
microfluidic system and barcoded beads. The library
of both full-length TCR/BCR sequences and mRNA
sequences can be constructed from a single sample,
followed by high-throughput sequencing and
bioinformatics analysis.
Investigate T/B cell heterogeneity, TCR/BCR diversity, antigen specificity, gene expression profiling, etc.
- Achieve efficient capture of V(D)J sequences with a mapping rate greater than 90%, ensuring more accurate data for immune research on immune cell diversity, clonal expansion, immune response, and vaccine development.
- Acquire complete coverage of the full-length V(D)J sequences, providing more comprehensive information for studying autoimmune and infectious diseases, elucidating the underlying pathogenesis, or exploring innovative therapeutic strategies.
- Provide accurate representation of α/β (light/heavy) chain pairing information of TCR or BCR. Pairing information from the same cell reveals the receptor-antigen specificity of immune cells.
- Simultaneous immune repertoire (BCR/TCR) and gene expression profiling from a single sample, linking cell types with comprehensive immunophenotyping.
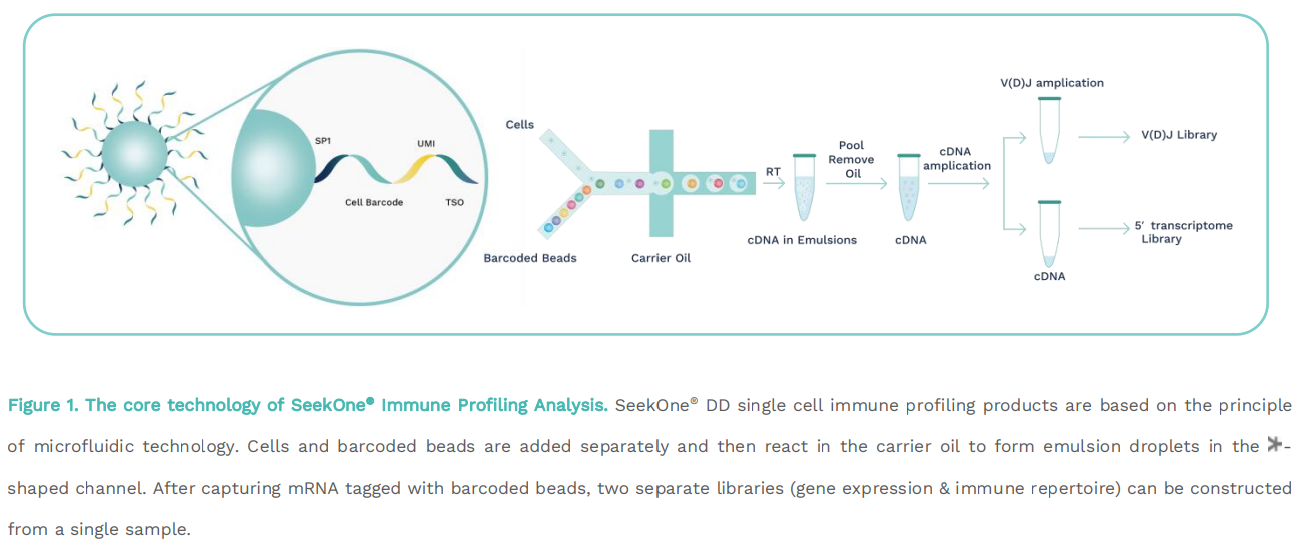
The core technology of SeekOne
®
Immune Profiling Analysis. SeekOne
® DD single cell immune profiling products are based on the principleof microfluidic technology. Cells and barcoded beads are added separately and then react in the carrier oil to form emulsion droplets in the-shaped channel. After capturing mRNA tagged with barcoded beads, two separate libraries (gene expression & immune repertoire) can be constructedfrom a single sample.
Product Features
Efficient Capture Efficiently capture V(D)J sequences with an alignment rate greater than 90%. Obtain more accurate and authentic capture data.
Full Length Complete coverage of the full-length V(D)J sequences. Analyze all V(D)J sequences, including CDR3 sequences, for more comprehensive information.
One sample, Two profiles Simultaneous profiling of immune repertoire (BCR/TCR) and 5’ gene expression Multiple libraries are constructed from a single sample, which is more economical and suitablefor samples with limited amounts, such as needle biopsy samples.
True Alignment
True representation of α/β (light/heavy) chain pairing information of TCR (BCR). Reports pairing information from the same cell to reveal receptor-antigen specificity of theimmune cells.
Product Specifications
- Up to 90% of reads mapped to any V(D)J gene
- Rapid generation of 150,000 water-in-oil droplets in 3 minutes
- Efficiently capture 500-12,000 cells per channel
- Flexible running of 1~8 samples in parallel
- Cell size flexibility: cell diameter of 5~40 μm
- High cell capture rates of up to 65%
- Low doublet rates of under 0.3% per 1,000 cells
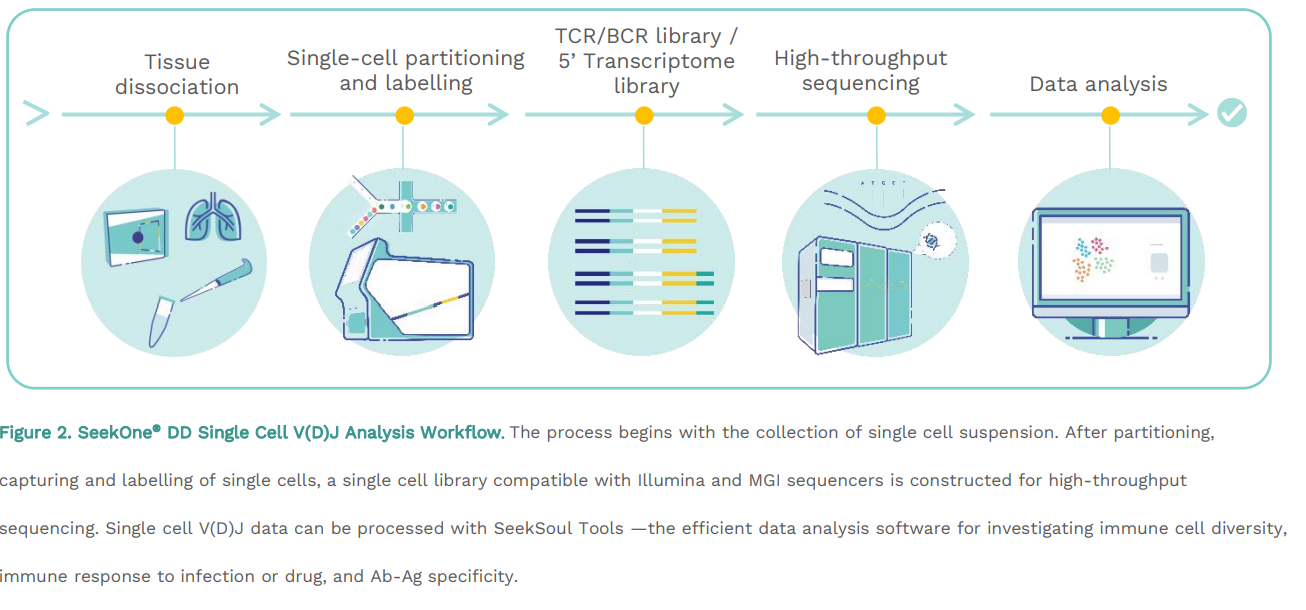
Workflow Steps
Tissue dissociation Single-cell partitioning and labelling
TCR/BCR library /
5’ Transcriptome
library
High-throughput sequencing Data analysis
Figure 2. SeekOne
® DD Single Cell V(D)J Analysis Workflow. The process begins with the collection of single cell suspension. After partitioning, capturing and labelling of single cells, a single cell library compatible with Illumina and MGI sequencers is constructed for high-throughput sequencing. Single cell V(D)J data can be processed with SeekSoul Tools —the efficient data analysis software for investigating immune cell diversity,
immune response to infection or drug, and Ab-Ag specificity