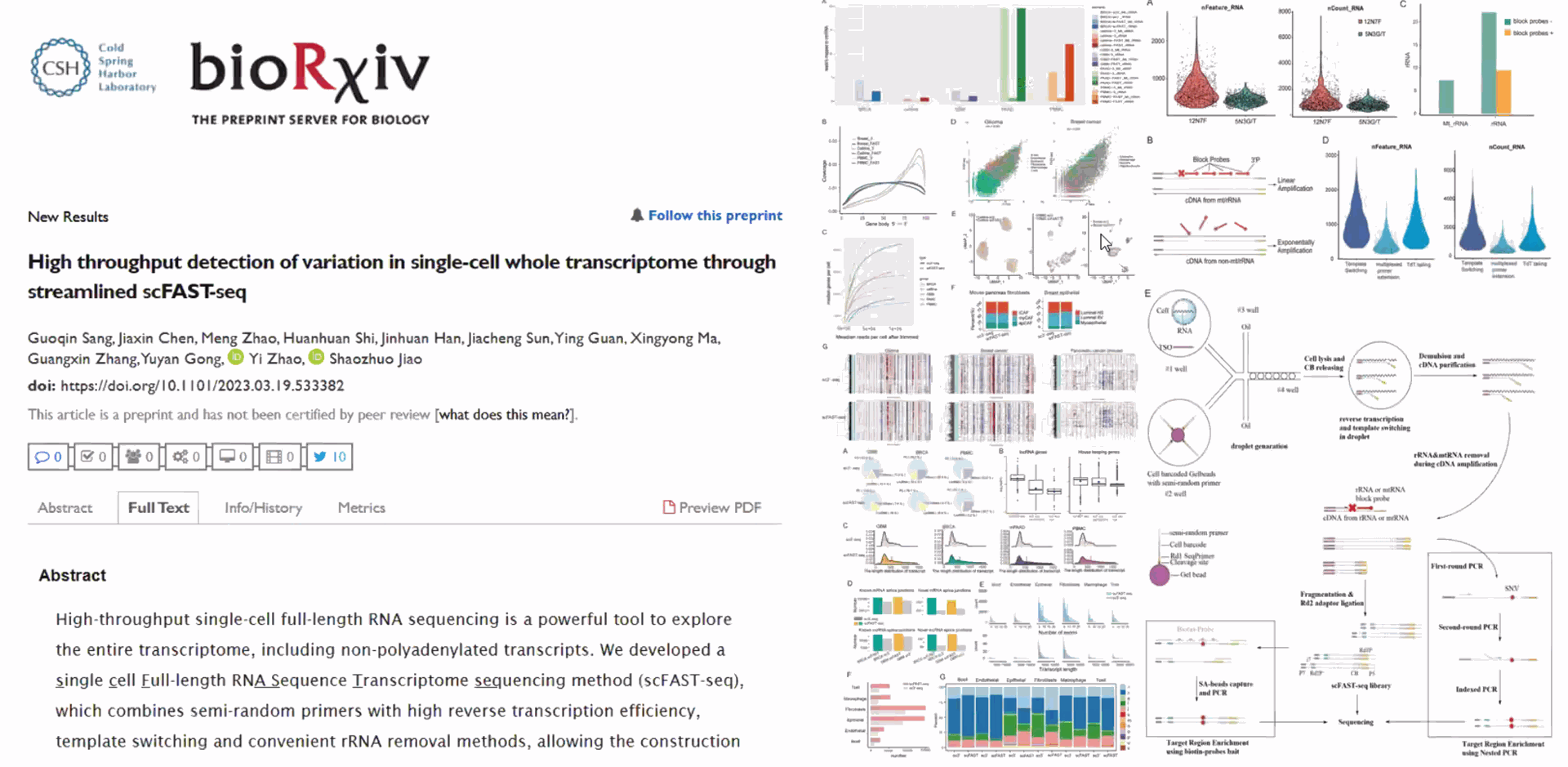
SeekOne Chromium comparative study
10x Genomics was compared to the SeekGene scRNA seq Seekone Instrument
Chromium Xo is a high-performance single cell scRNA seq
It uses GEM-X technology for 3’ whole transcriptome profiling, Chromium Xo.
Why Seekgene's Single Cell 3' Transcriptome kit called scFAST seq is better?

—— Investigate gene expression with accuracy and efficiency
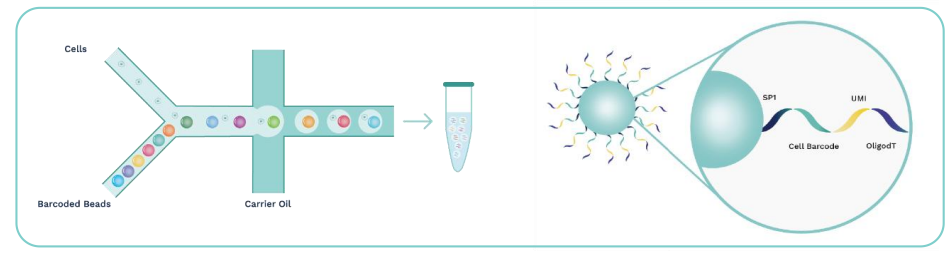
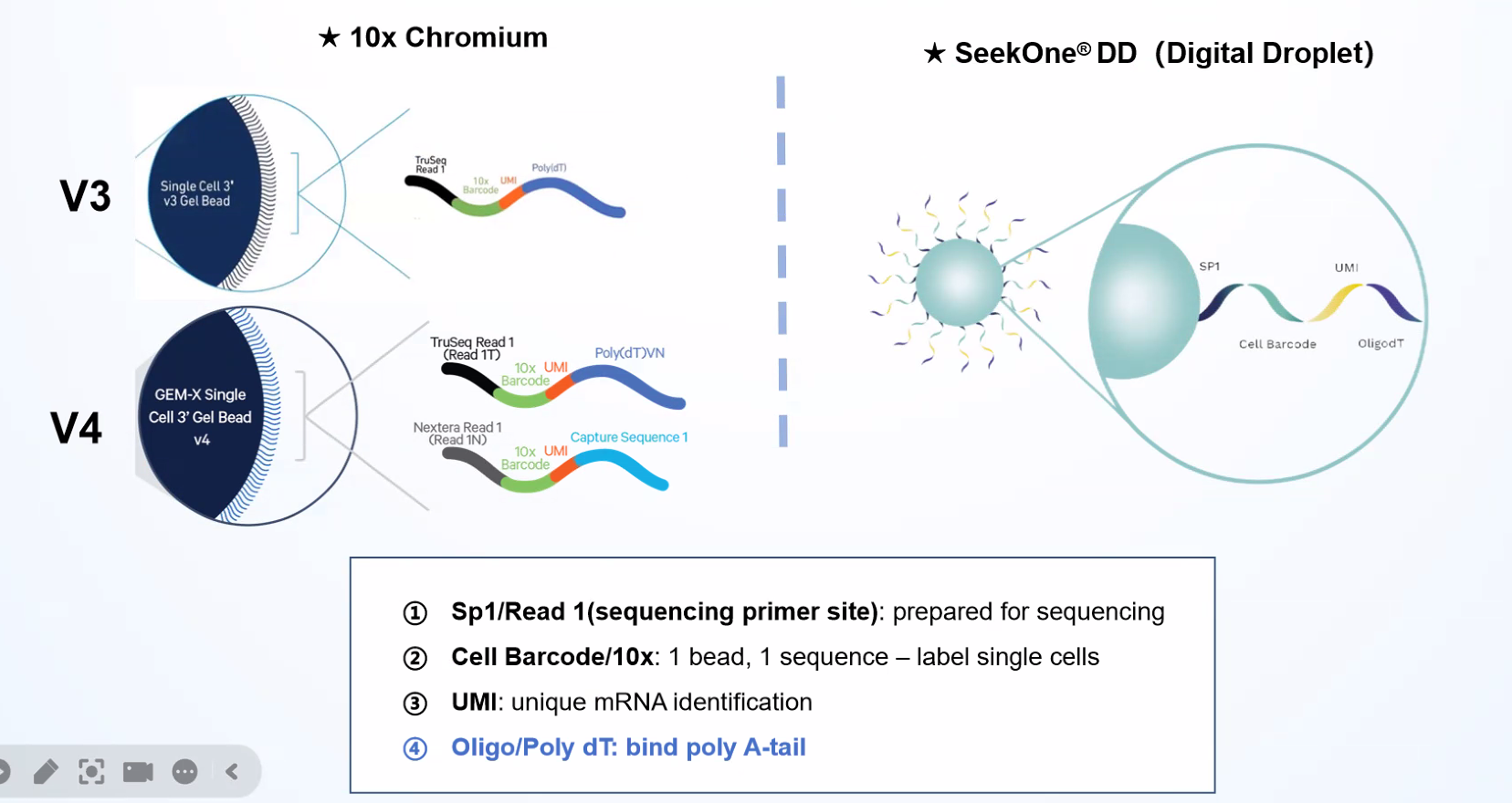
SeekOneDD (Digital Droplet) Single Cell 3' Transcriptome-seq is a high-throughput single-cell RNA-seq library kit achieves single-cell partitioning and labelling by the microfluidic system and barcoded beads. The kit needs to be used with the SeekOne
Digital Droplet System (SeekOne
DD) to complete the whole process from single-cell transcriptome barcoding to library construction.

SeekOneDD single cell 3' transcriptome kit includes: SeekOne
DD Chip S3 (Chip S3 for short), Gasket, Carrier Oil, SeekOne
DD 3' Barcoded Beads (Barcoded Beads for short), amplification reagents, library construction reagents, and single cell data analysis software (SeekSoul
Tools).
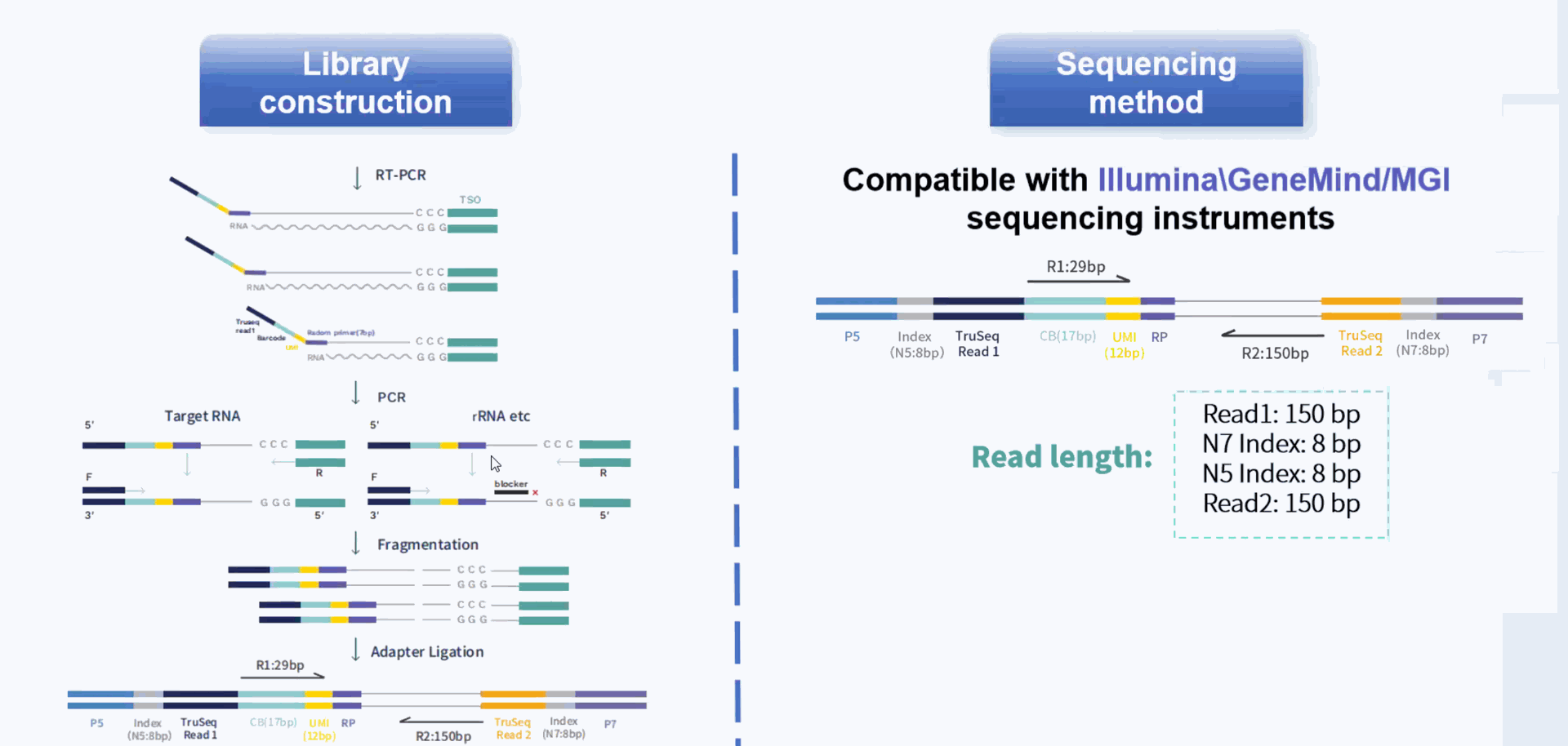
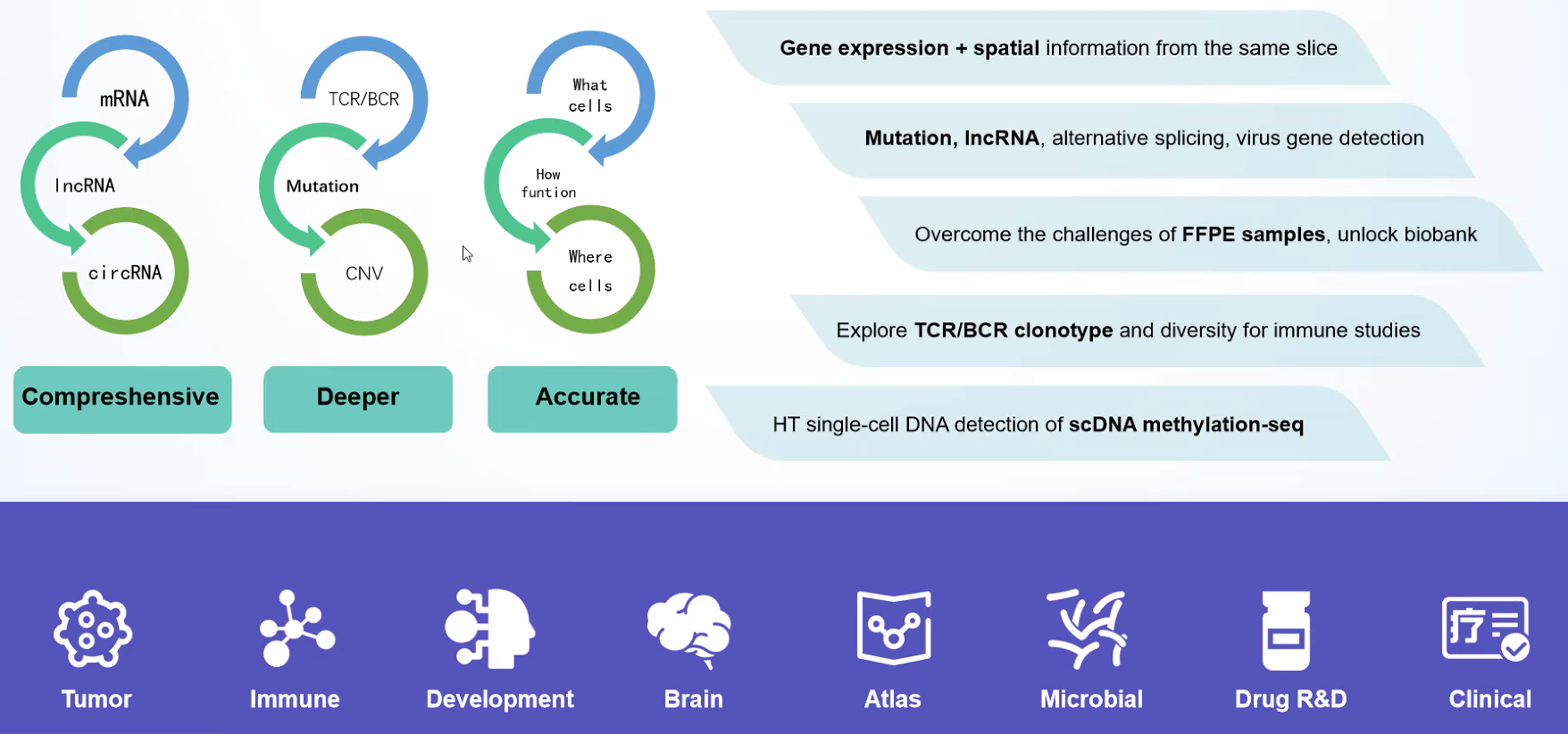
Based on the principle of microfluidic technology, the kit uses emulsion droplets to separate and capture individual cells, and then recruits nucleic acid-modified Barcoded Beads to molecularly label RNA from different cell sources. Finally, a high-throughput single-cell transcriptome library compatible with Illumina and MGI sequencing system will be constructed, which can be used for research on tumors, immunity, cell development, viral infections, drug discovery, and target screening, etc.
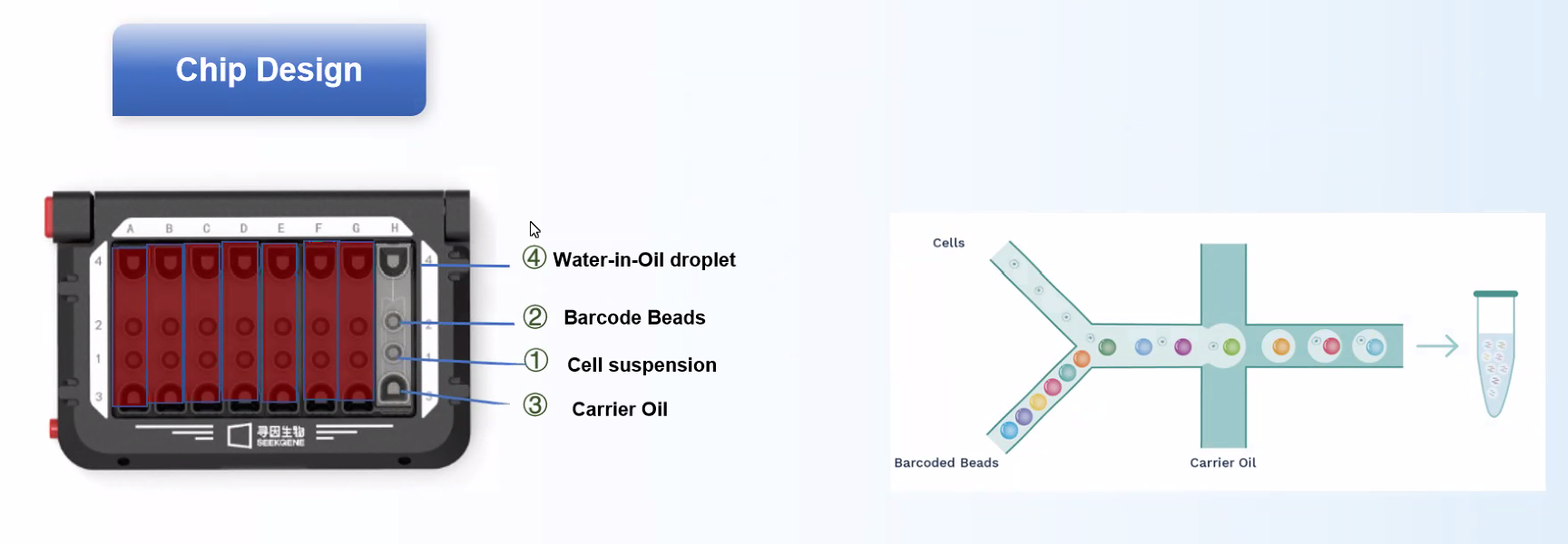
1. Seekgene has better Single cell capture and labelling
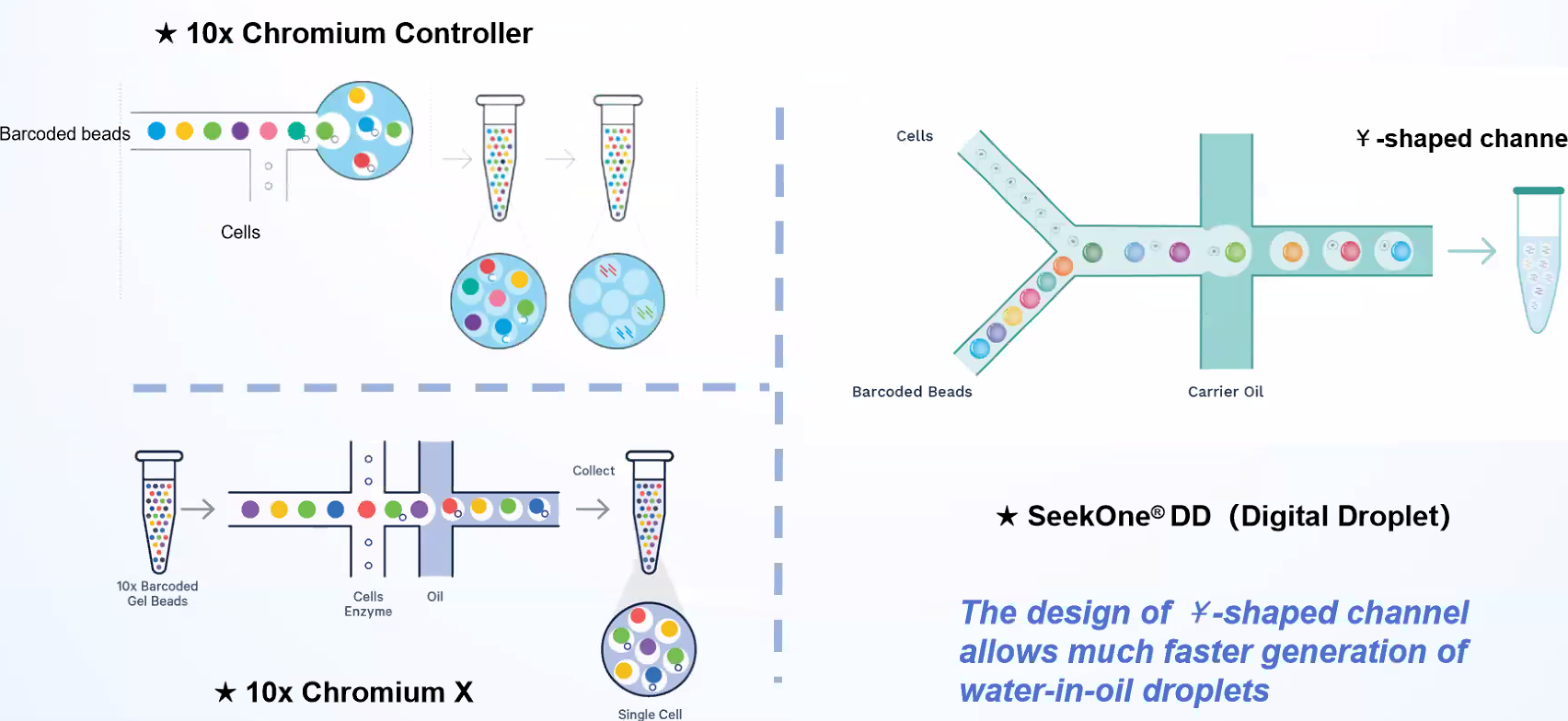
Cells and barcoded beads are added separately and then react in the carrier oil to form emulsion droplets in the -shaped channel. After that, mRNA molecules released by the cells are captured by oligo(dT) on the barcoded beads.

- Workflow
- Product Features
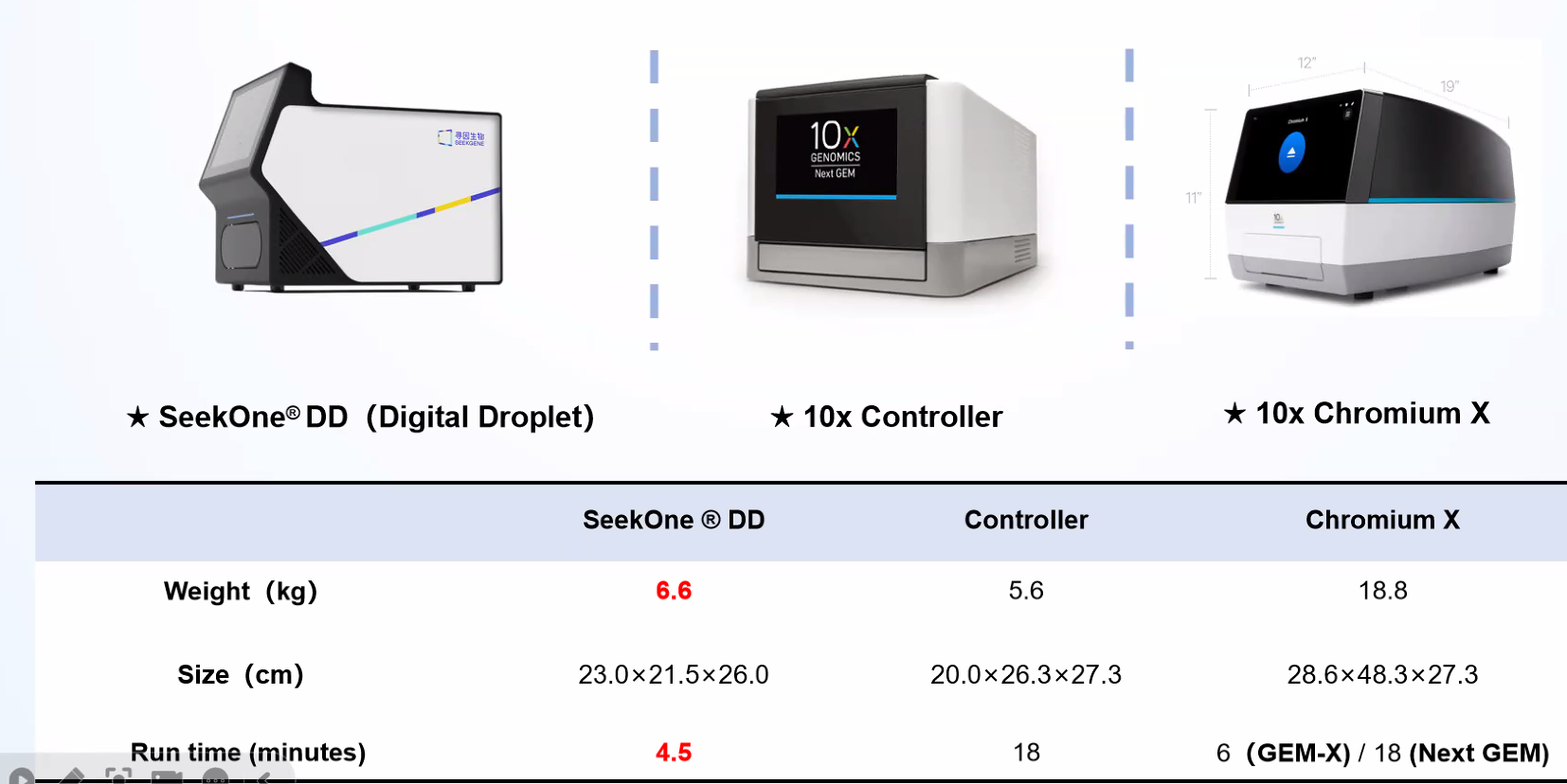
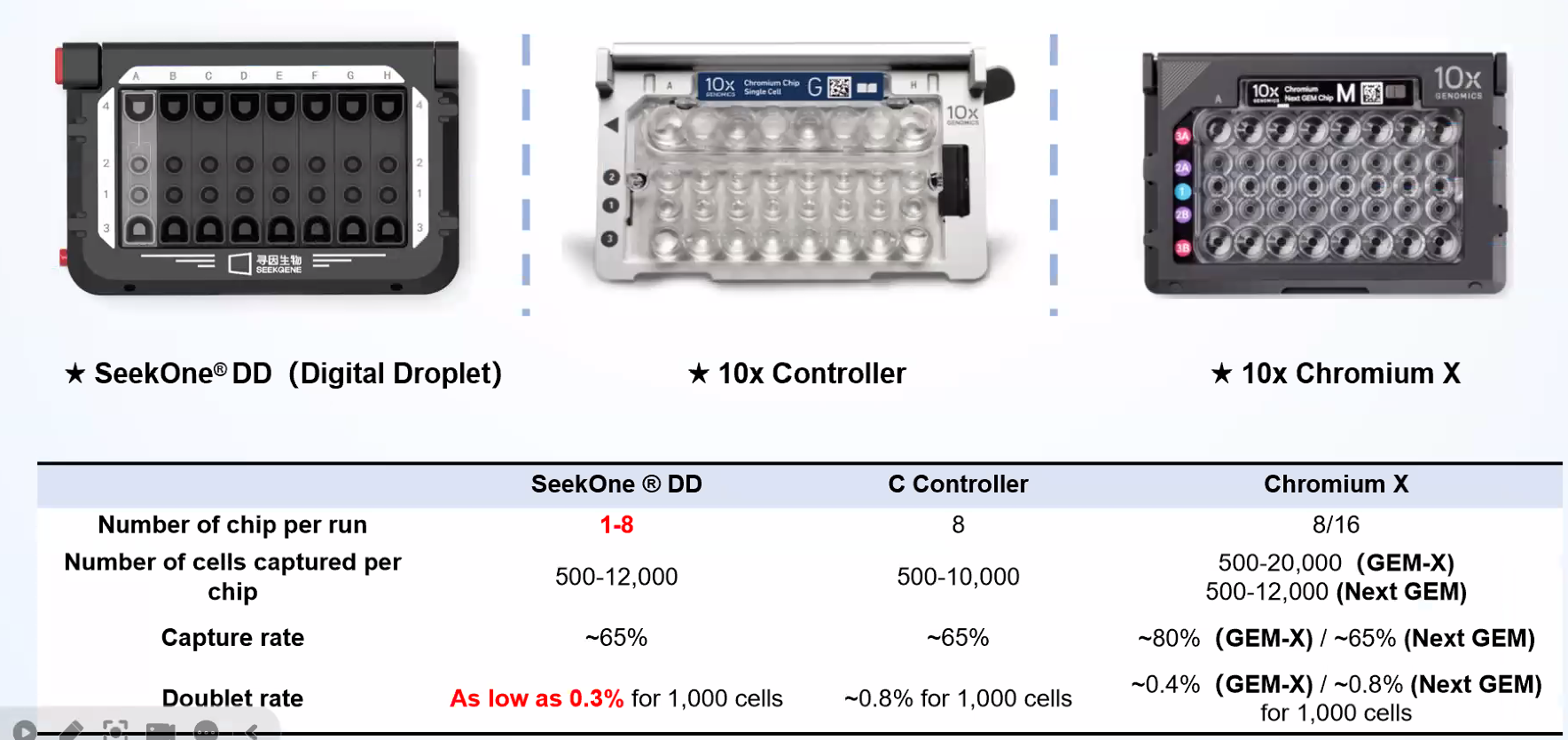
Compare Compatible Instrument
SeekOneDD is much liter and better pricing than Chromium X
Lieven Gevaert, Bio-ir at Gentaur Institute Leuven Belgium
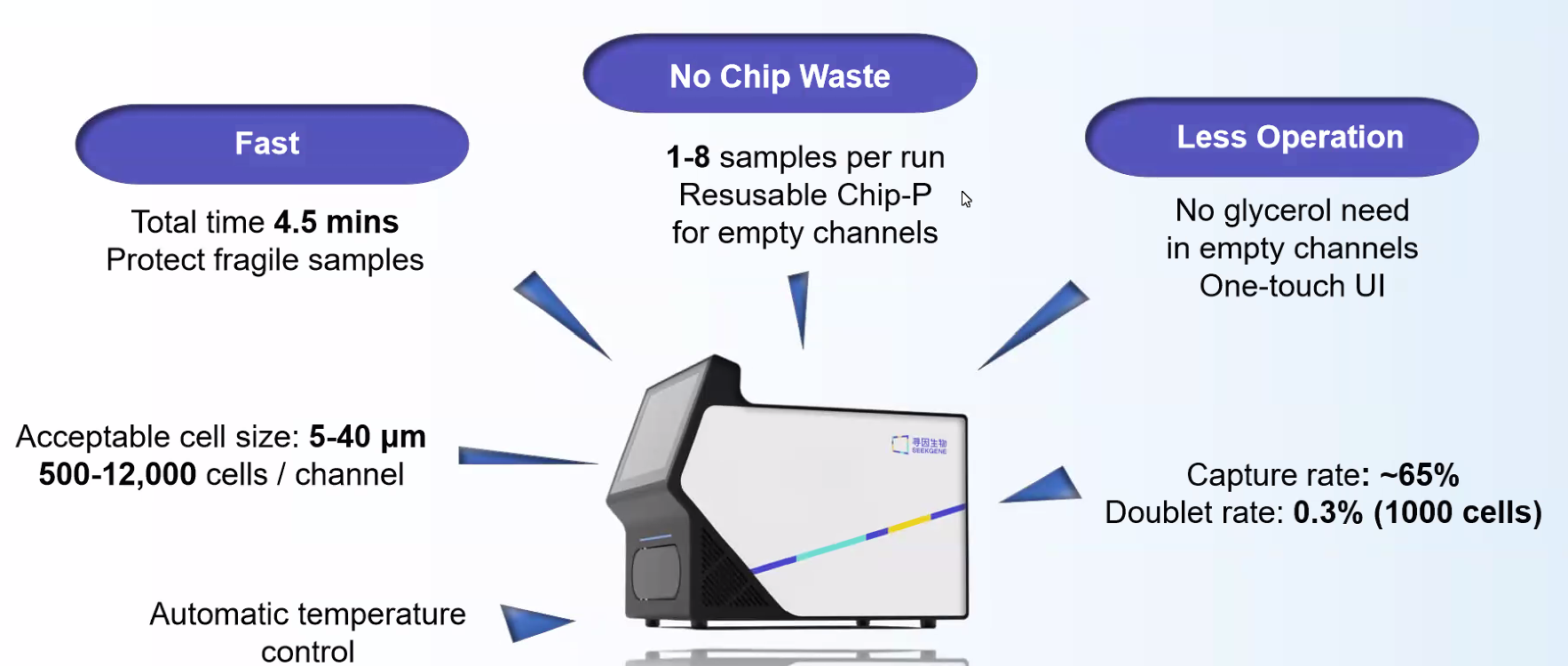
1-8samples can be flexibly run at a time
500-12,000 cells can be captured with a single channe 150,000
emulsion droplets can be generated in 3 minutes
Recovery of ~1000 cells with
doublet rates of
~0.3% per 1,000 cells
2. Seekgene has an Unlimited Library Construction
● Compatible
Compatible with single cell 3' transcriptome sequencing (3' scRNA-seq) and single nucleus RNA sequencing (snRNA-seq). Dual index libraries compatible with both Illumina and MGI sequencers.
● Seekgene is Faster than 10x
Rapid generation of 150,000 water-in-oil droplets in 3 minutes with high success
rates, especially for precious and sensitive samples.
● Seekgene is more Cost-saving compared to 10x Chromium
Unique place chip design eliminates chip waste, reducing operation time and cost
● Suitable sample types
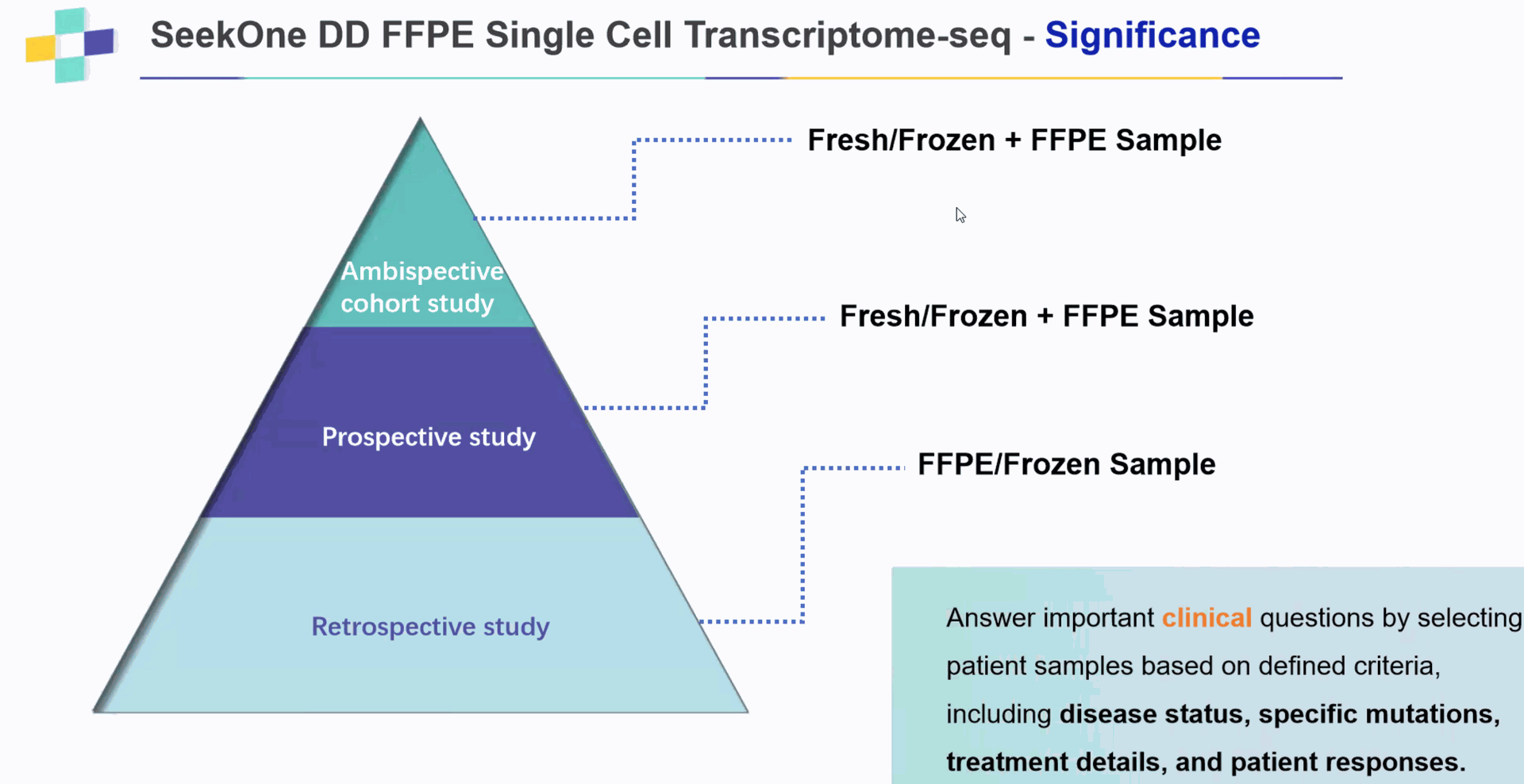
a. Delicate samples with low cell viability:brain tissue, retinal tissue, all tissues from aged individuals
b. Tissues requiring nuclei isolation: frozen tissues, pancreatic tissues, neurons, adipose tissue, etc.
c. Larger cohort or atlas studies where multiple samples need to be processed simultaneously.

SeekOneDigital Droplet System
Reverse transcription
Template conversion and extension
Fragmentation and end repair
connector connection
library amplification

SeekOneDD
Single Cell 3' Transcriptome kit
SeekOneDigital Droplet System
Seekgene has better Reagents
More capable Instrument
Seekgen has better Software
SeekOneDD

Single Cell Immune Profiling Kit
SeekOneDD
Single Cell Full-length RNA Sequence Transcriptome-seq kit
cDNA amplification
High cell capture rates of up to 65%
Order Information
Product Name | Cat.No. | Specification |
SeekOne® DD Single Cell 3’ Transcriptome-seq Kit |
in Gentaur we work to make single cell more accessible at lower costs, Chromium Xo is just the start. New Chromium innovations are coming soon, continuing our commitment to provide superior performance over alternative methods and reduce budget constraints at every step. What does that look like in your lab? Find out below.
The next generation of single cell is here
Introducing Chromium Single Cell Gene Expression v4 and Single Cell Immune Profiling v3, powered by new GEM-X technology
- Partition and barcode your cells 3x faster
- Achieve cell recovery efficiency rates of up to 80%
- Reduce multiplet rates two-fold
- Lower sequencing costs by up to 50%
Uncover new insights
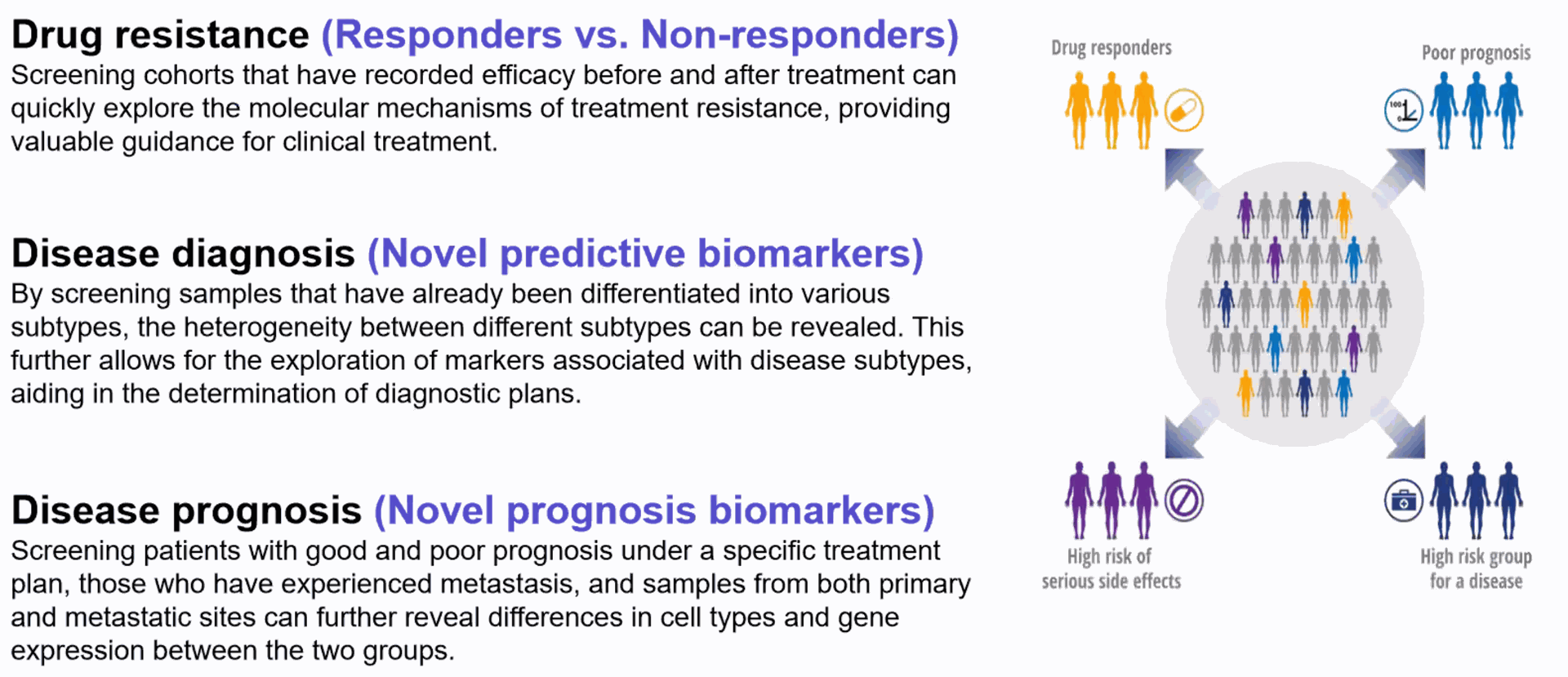
Discover cellular diversity, rare cells, and disease-linked cell types.
Single cell immune profiling fuels biomarker discovery in cancer samples
Check out three separate studies from researchers at Columbia University and MD Anderson that provide impactful insights into cancer research.
Rpostead blog post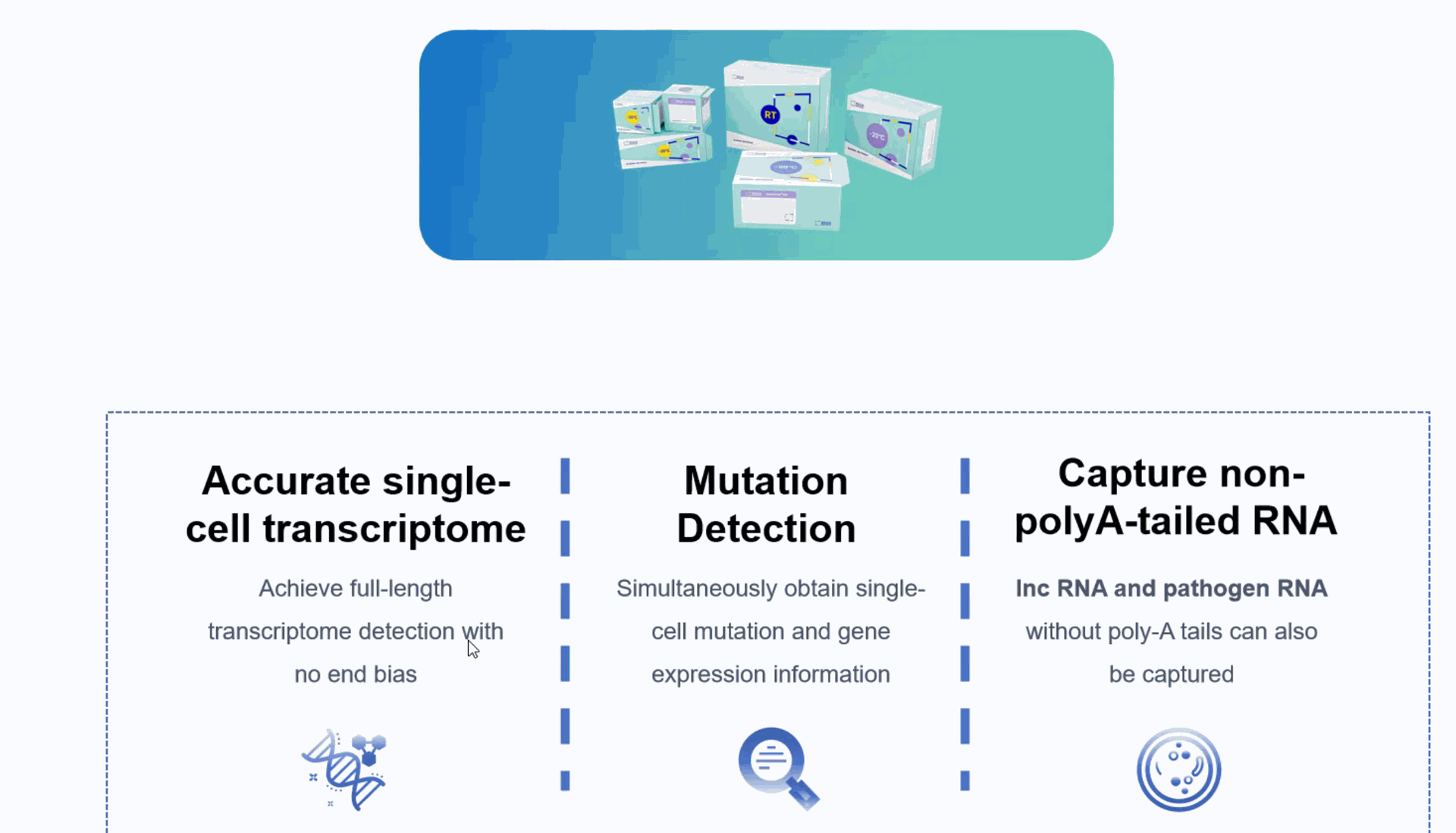
Single cell multiomics reveals kidney cell behind erythropoietin, master regulator of red blood cell production
Explore a recent Nature Medicine publication, where researchers used single cell transcriptomic and epigenetic analyses to identify the cell type behind the production of erythropoietin, opening up anemia treatment possibilities.
Organoids bring drug discovery and development to the culture hood
Explore the work of four research teams using single cell RNA-seq and organoids to identify druggable pathways in liver disease, screen drugs for kidney disease, model drug metabolism in the liver, and study therapeutic responses in oncology patient-derived models.
Capture RNA, protein, chromatin and more
Get a complete picture with versatile assays that profile multiple analytes.
Compare single cell assaysAll (5)Gene Expression (4)B-cell or T-cell receptors (1)Chromatin accessibility (2)Cell surface protein (3)
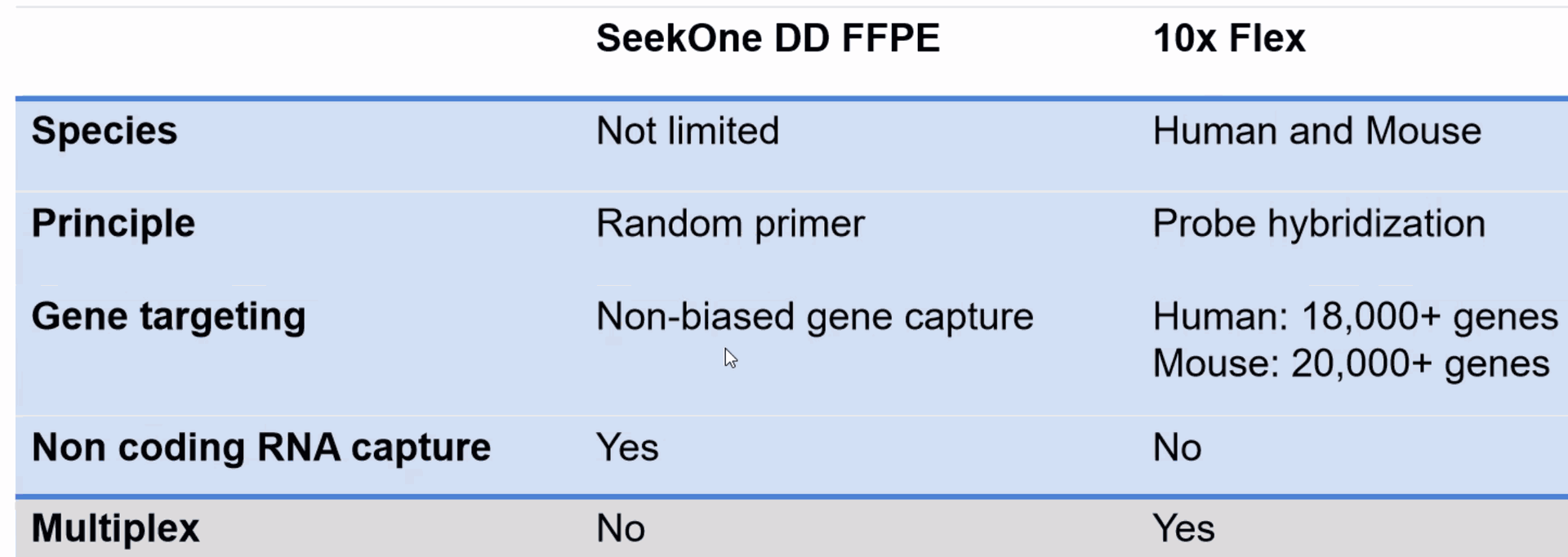
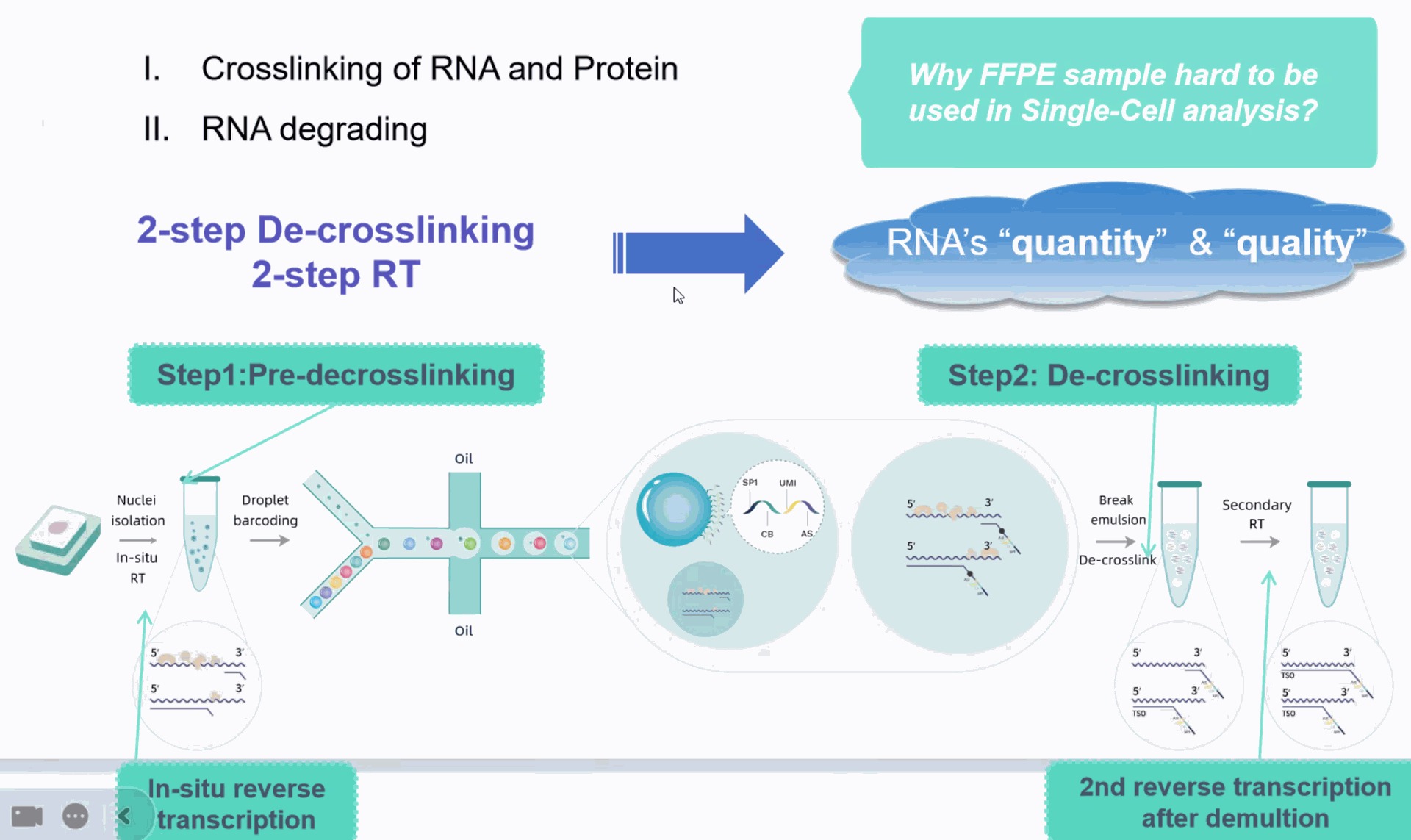
Probe-based gene expression for fresh, frozen, and FFPE samples
What is a FFPE sample?![]()
Formalin-Fixed Paraffin-Embedded sample
Formalin-Fixed Paraffin-Embedded (FFPE) tissue specimens have been a staple of research and therapeutic applications for decades. FFPE is a form of preservation and preparation for biopsy specimens that aids in examination, experimental research, and diagnostic/drug development.
Our most sensitive whole transcriptome analysis with the flexibility to fix, batch, and run samples on your schedule.
Single Cell Gene Expression Flex
3' gene expression
A versatile solution providing 3' transcriptomic profiling with options for multiomic analysis.
5' gene expression with immune repertoire profiling
Assay with multiomic capabilities and options for comprehensive analysis of immune diversity, including BCR or TCR repertoire profiling.
Chromatin accessibility and gene expression from the same nucleus
Simultaneously profile the transcriptome and epigenome at single cell resolution.
Chromatin accessibility
Analyze thousands of unique open chromatin fragments per cell with scATAC-seq.
Single Cell ATACAdditional capabilitiesAll (3)Compatible with Gene Expression (3') (2)Compatible with Immune Profiling (5') (3)
Directly link CRISPR guide RNAs to the resulting perturbed phenotypes
Capture protein alongside gene expression to better characterize complex biology
Accelerate therapeutic discovery with rapid, antigen-specific clonotype identification
Barcode Enabled Antigen Mapping
Get the instrument-supported Chromium advantage
- Automate the most critical steps for more reproducible results and a lower risk of manual error than with instrument-less methods
- Ensure high success rates and consistent performance, with a demonstrated R2 value of 0.97 across runs, users, and timepoints
- Partition tens of thousands of cells in mere minutes, saving on time and labor costs with 1-day workflows and just 3–4 hours of hands-on time
- Use our intuitive push-button interface for efficient cell processing without the hassle of cumbersome pipetting methods
PRODUCTS
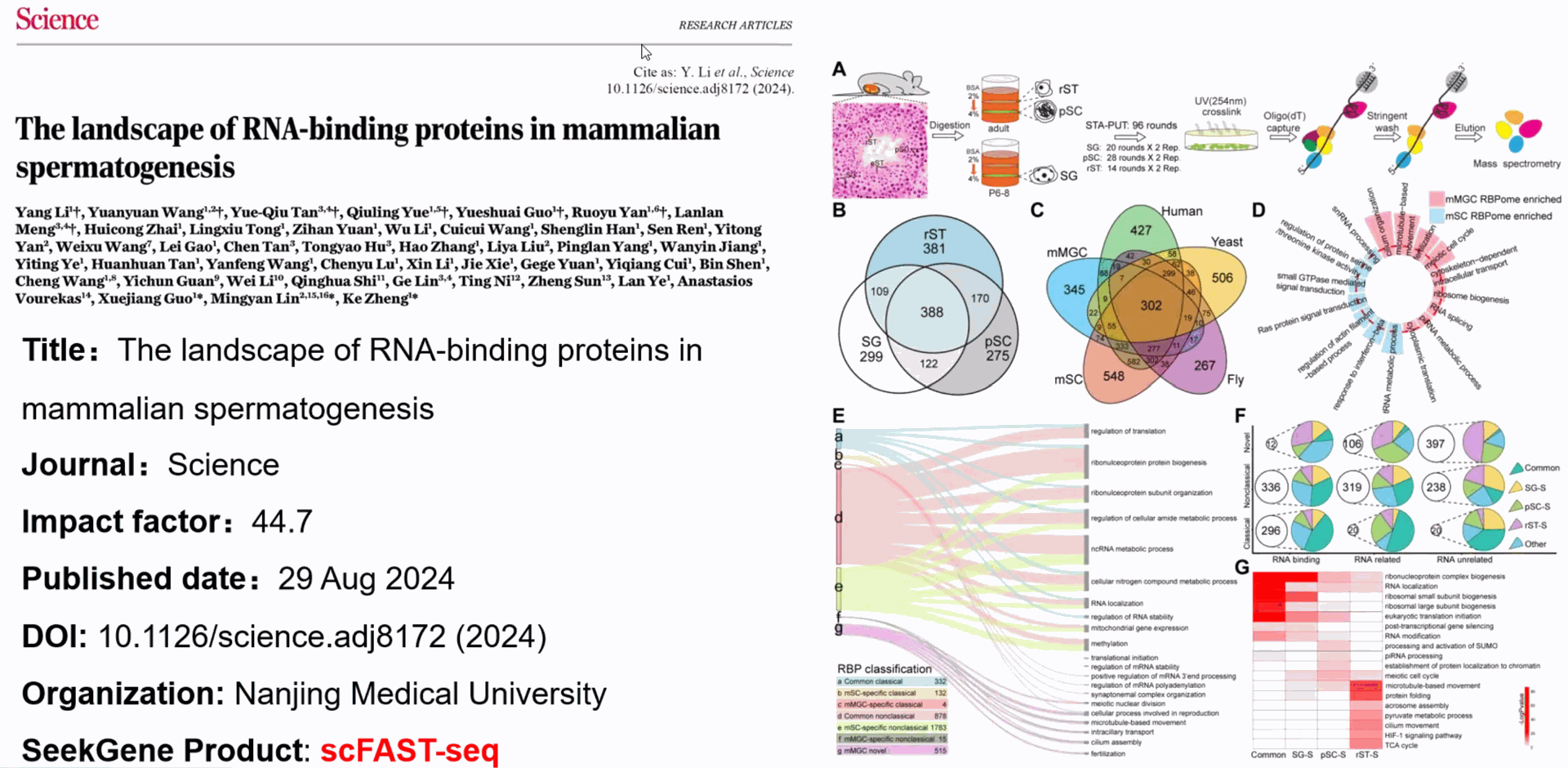
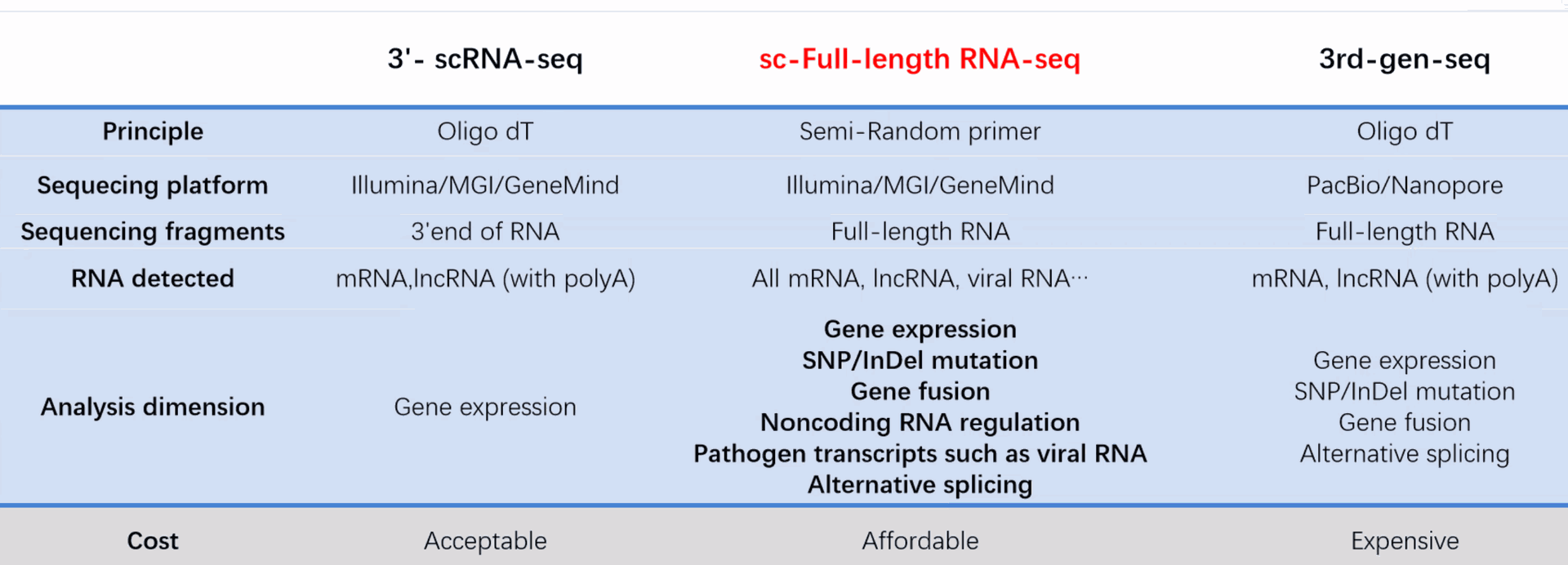
Single Cell Full-length RNA Sequence Transcriptome-seq kit
—— The Time Has Come for A New Generation of Single Cell Transcriptome Profiling
SeekOneDigital Droplet (SeekOne
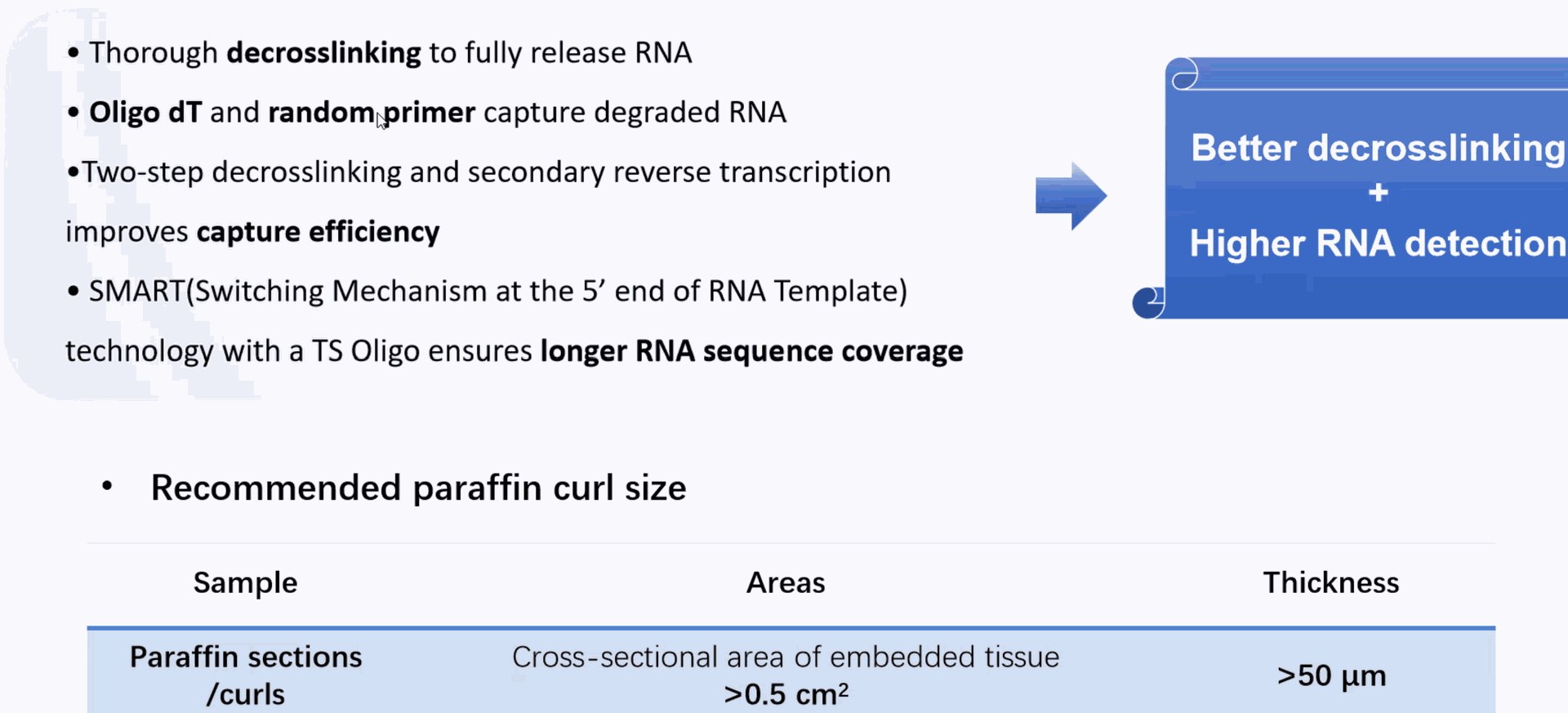
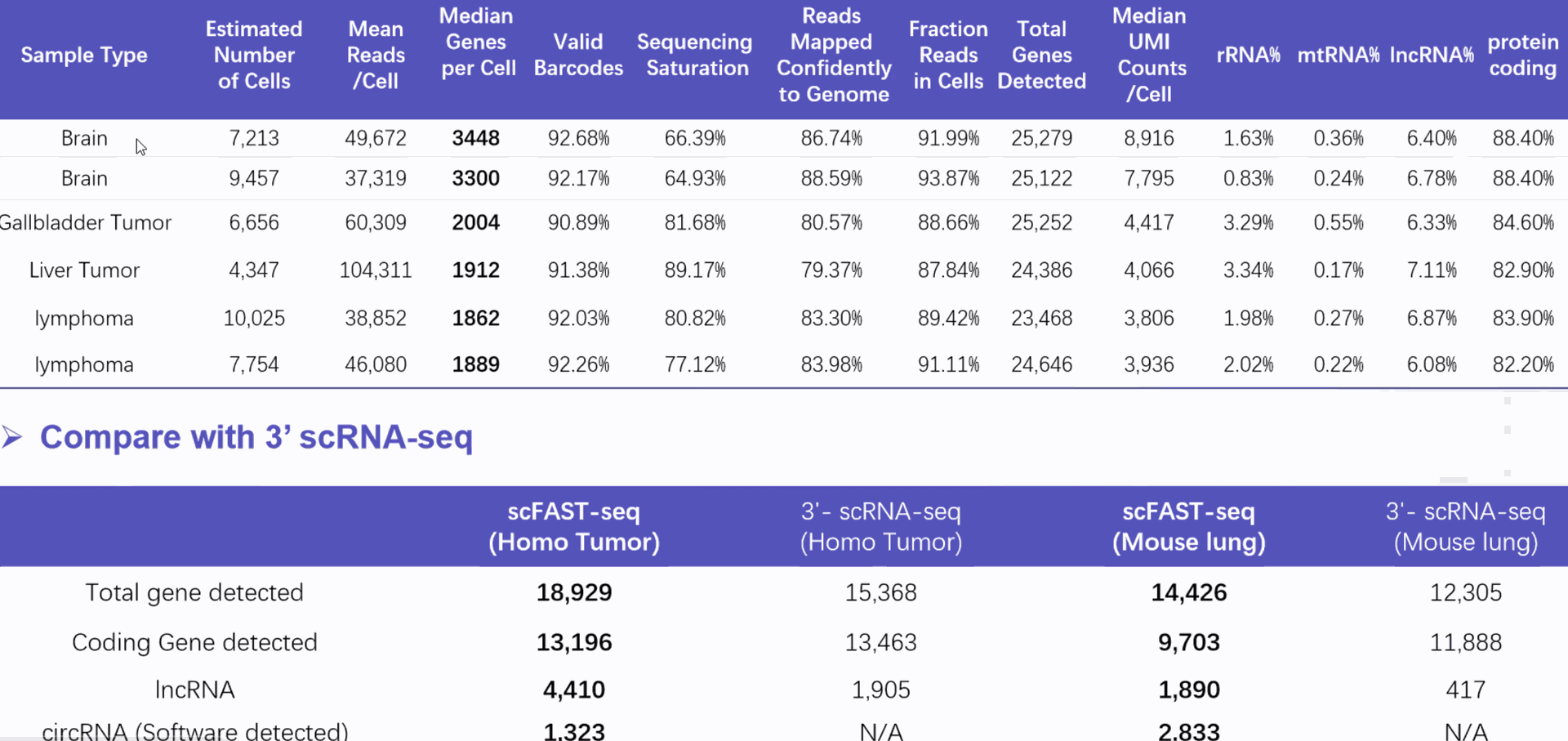
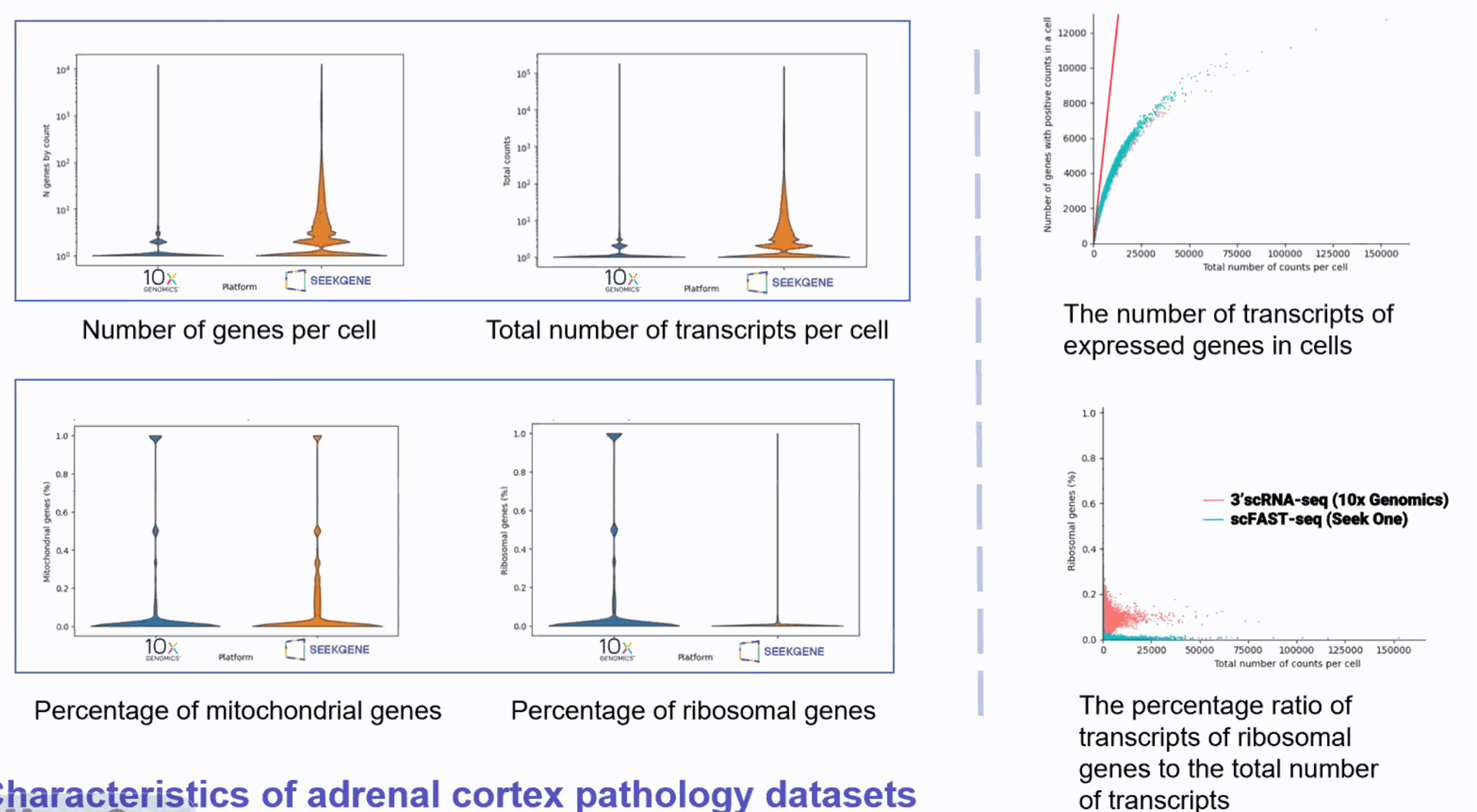
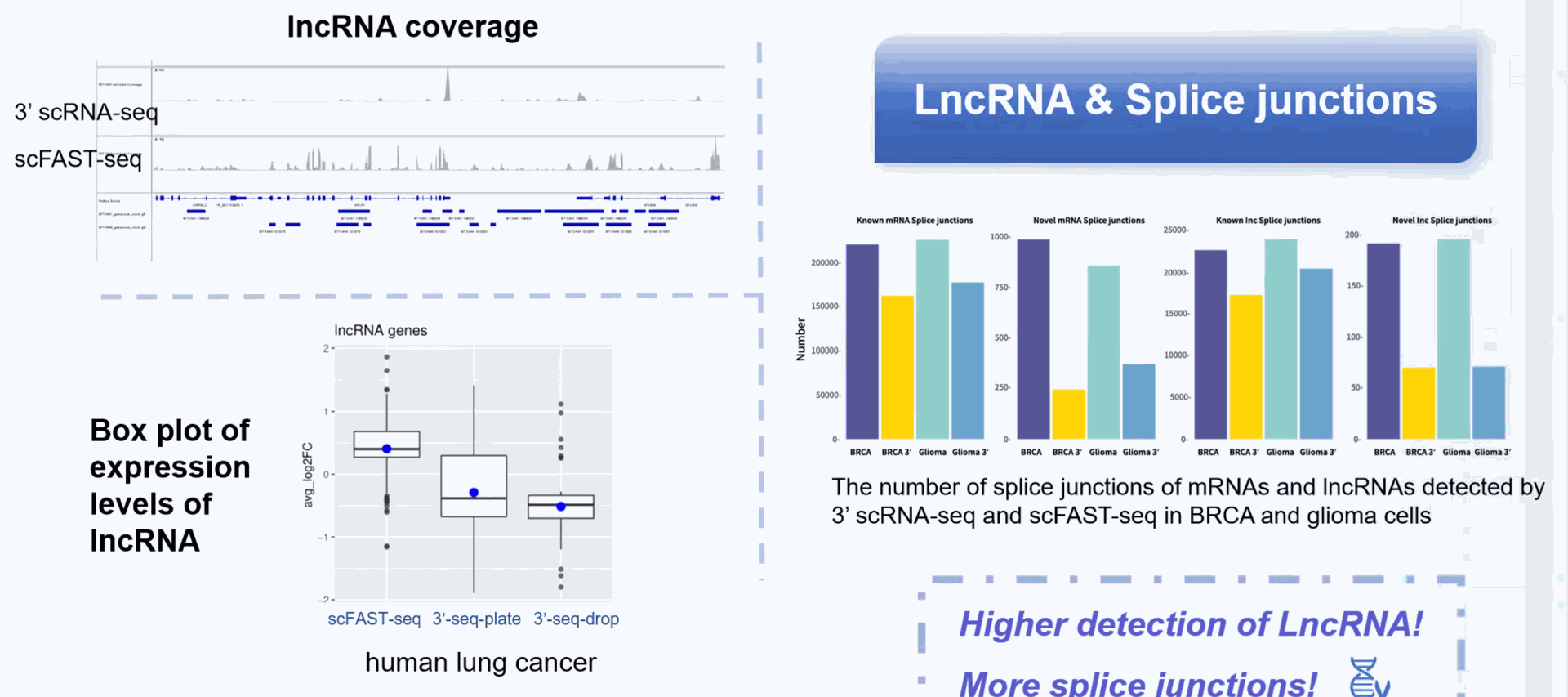
DD) High throughput Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit, self- developed by Beijing SeekGene BioSciences Co., Ltd., is a powerful commercial tool for high-throughput whole transcriptome profiling. Compared to conventional 3' scRNA-seq, scFAST- seq has distinct advantages in detecting non- polyadenylated transcripts, transcript coverage length, and identification of more splice junctions. With target region enrichment, scFAST-seq can simultaneously detect somatic mutations and cell states in individual tumour cells, providing valuable information for precision medicine.
SeekOneDD Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit includes SeekOne
DD Chip S3 (Chip S3), Gasket, Carrier Oil, SeekOne
DD scFAST-seq Barcoded Beads, amplification reagents, library construction reagents.
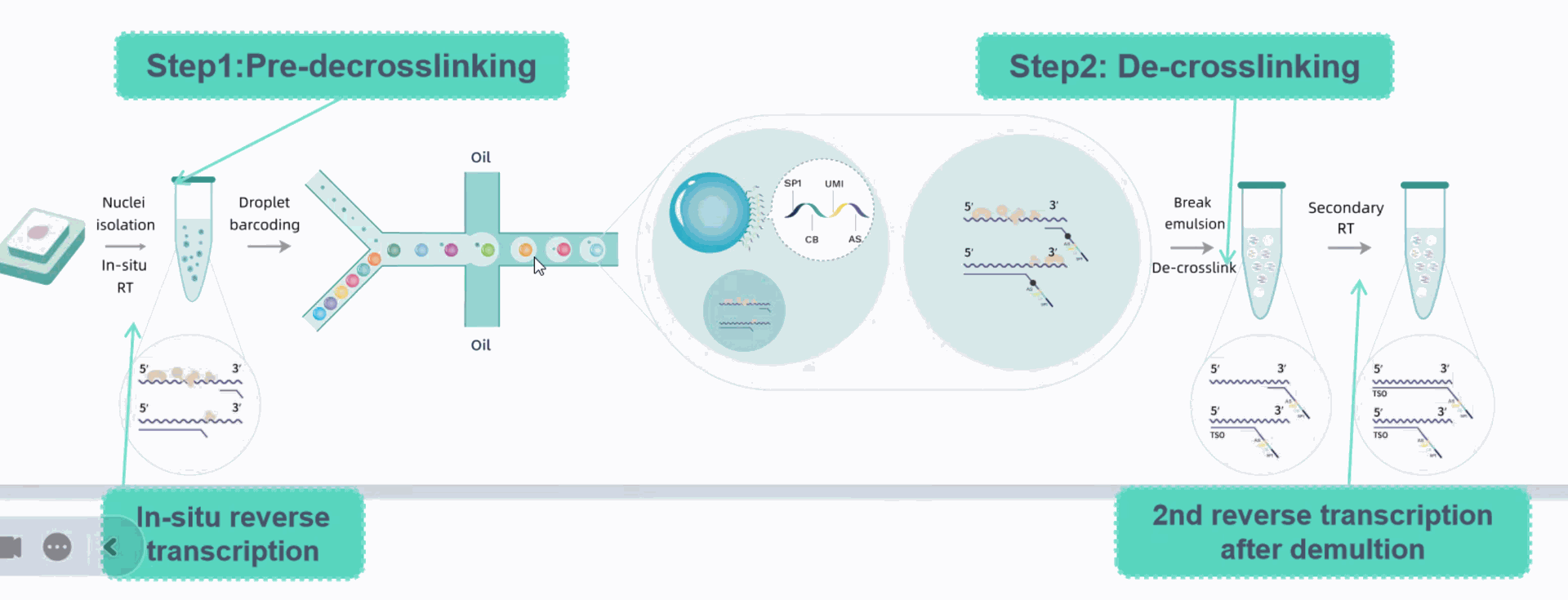
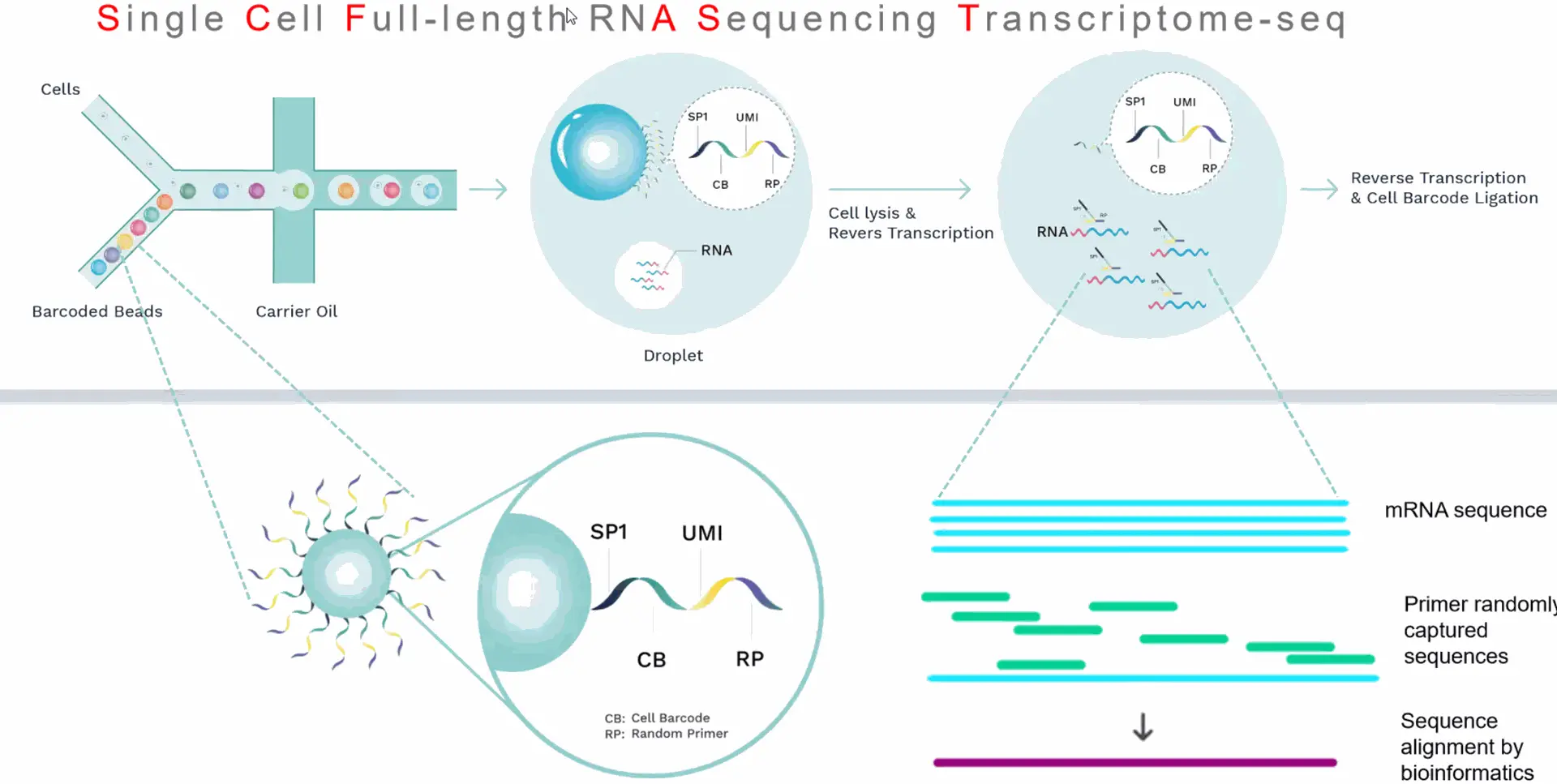
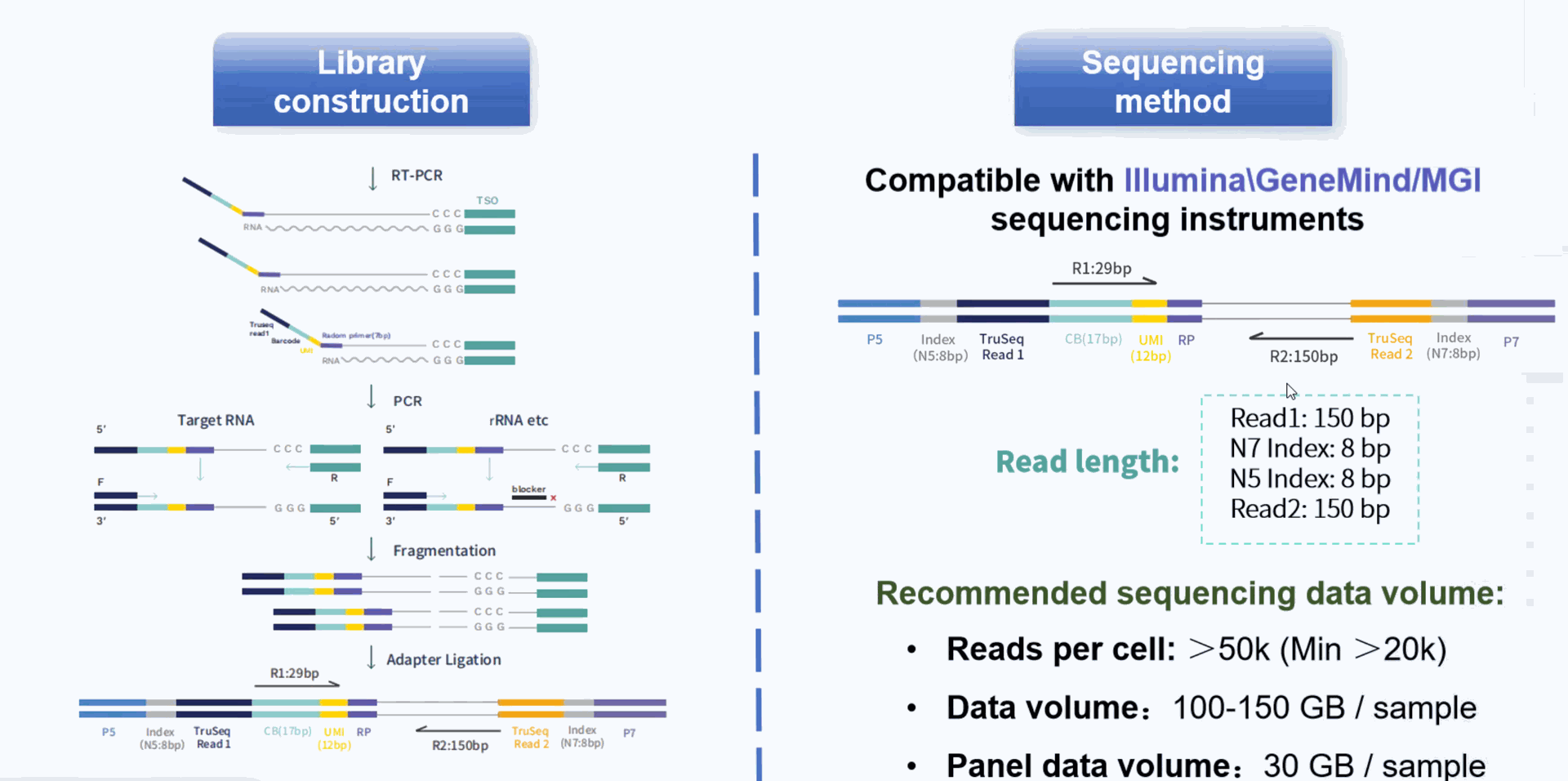
Using microfluidic digital droplets and Barcoded Beads coupled with random primers, single-cell partition and random capture of the whole transcriptome can be achieved in a step of generating water-in-oil droplets. This convenient process requires only a small amount of sample. The inclusion of an rRNA blocker enables efficient depletion of rRNA, delivering higher proportion of effective data.
Core technology and workflow
Compatible Instrument
SeekOneDD
SeekOneDigital Droplet System
SeekOneDD
Single Cell 3'Transcriptome-seq kit
Reagent
Instrument
Software
SeekOneDD
Single Cell Immune Profiling Kit
SeekOneDD
Single Cell Full-length RNA Sequence Transcriptome-seq kit
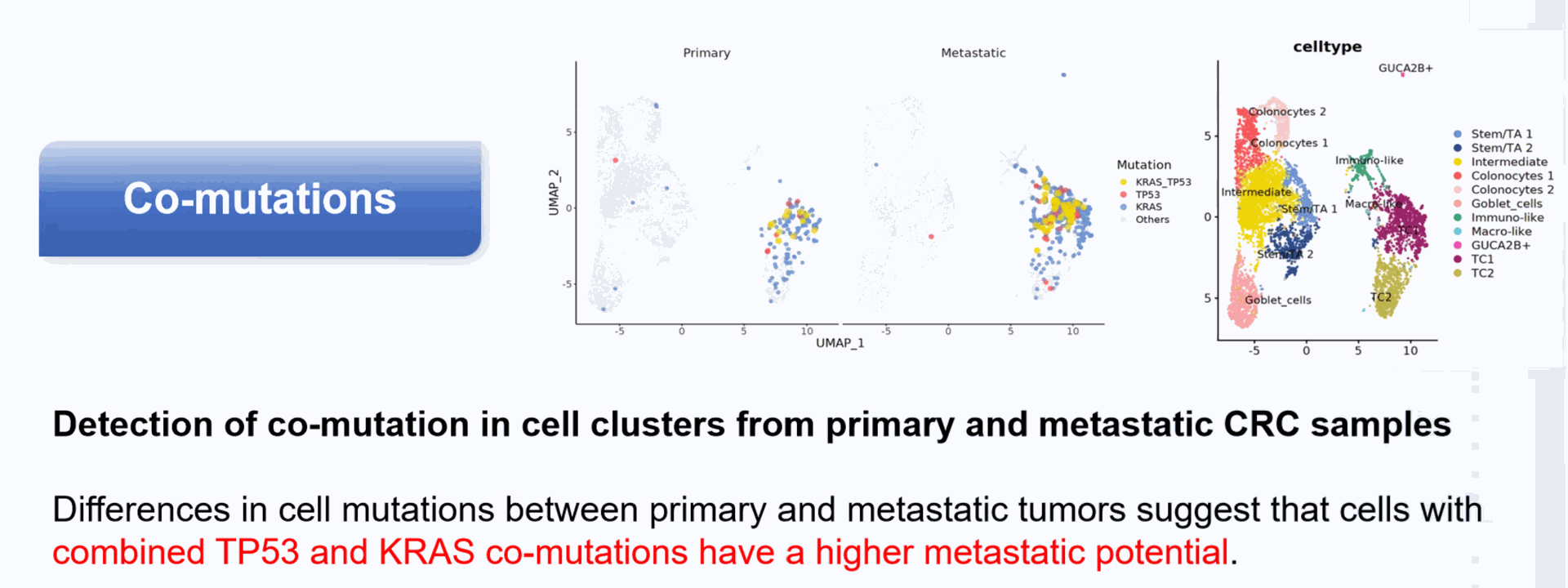
Detection of single cell mutation
Order Information
1-8 samples can be
flexibly run at a time
500-12,000 cells can be captured with a single channel
150,000 emulsion droplets can be generated in 3 minutes
Recovery of ~1000 cells with doublet rates of ~0.3% per 1,000 cells
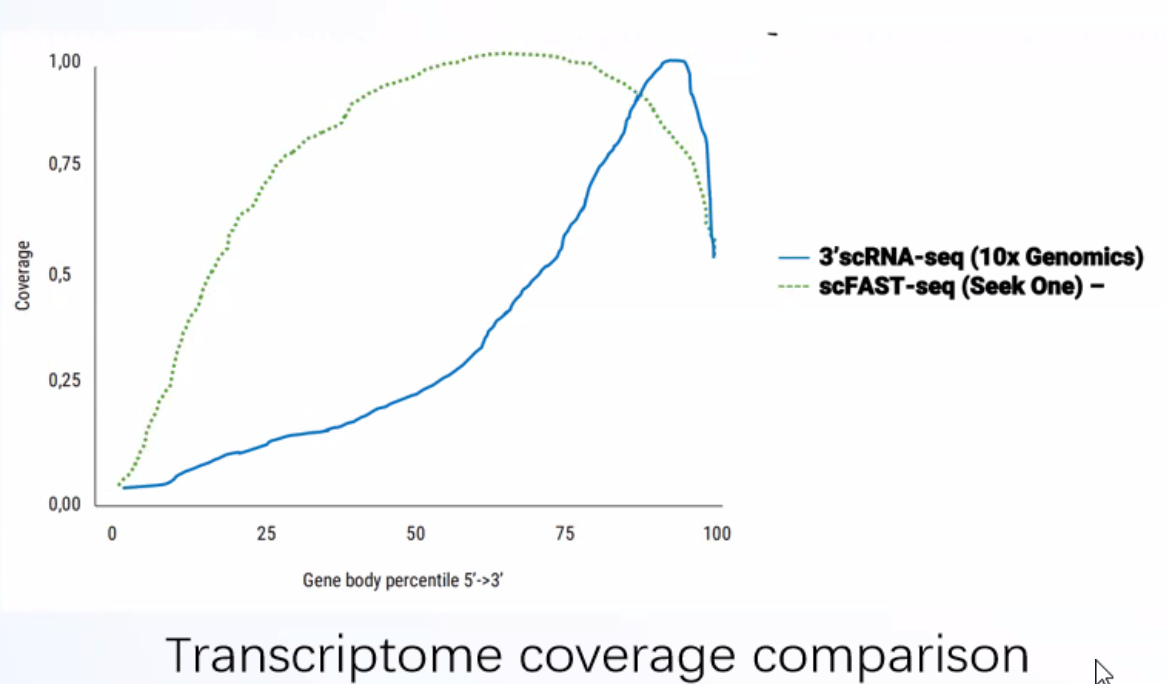
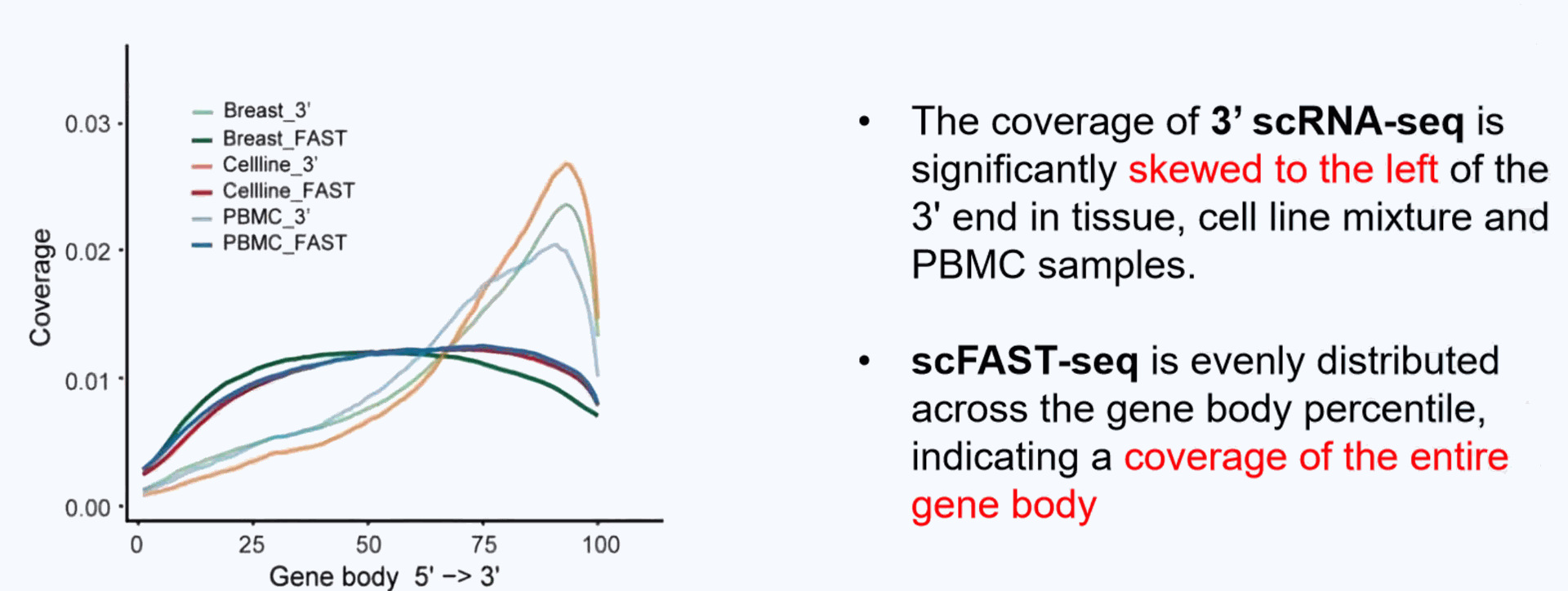
Even coverage of the full-length of the gene from 5' to 3'end
Seekgene Product Features
Seekgene More Accurate single-cell transcriptome: Achieve full-length transcriptome detection without being limited by the 3' or 5' end.
Genotype to phenotype:: Simultaneously obtain single-cell mutation and gene expression information, establishing the relationship between genotype and phenotype.
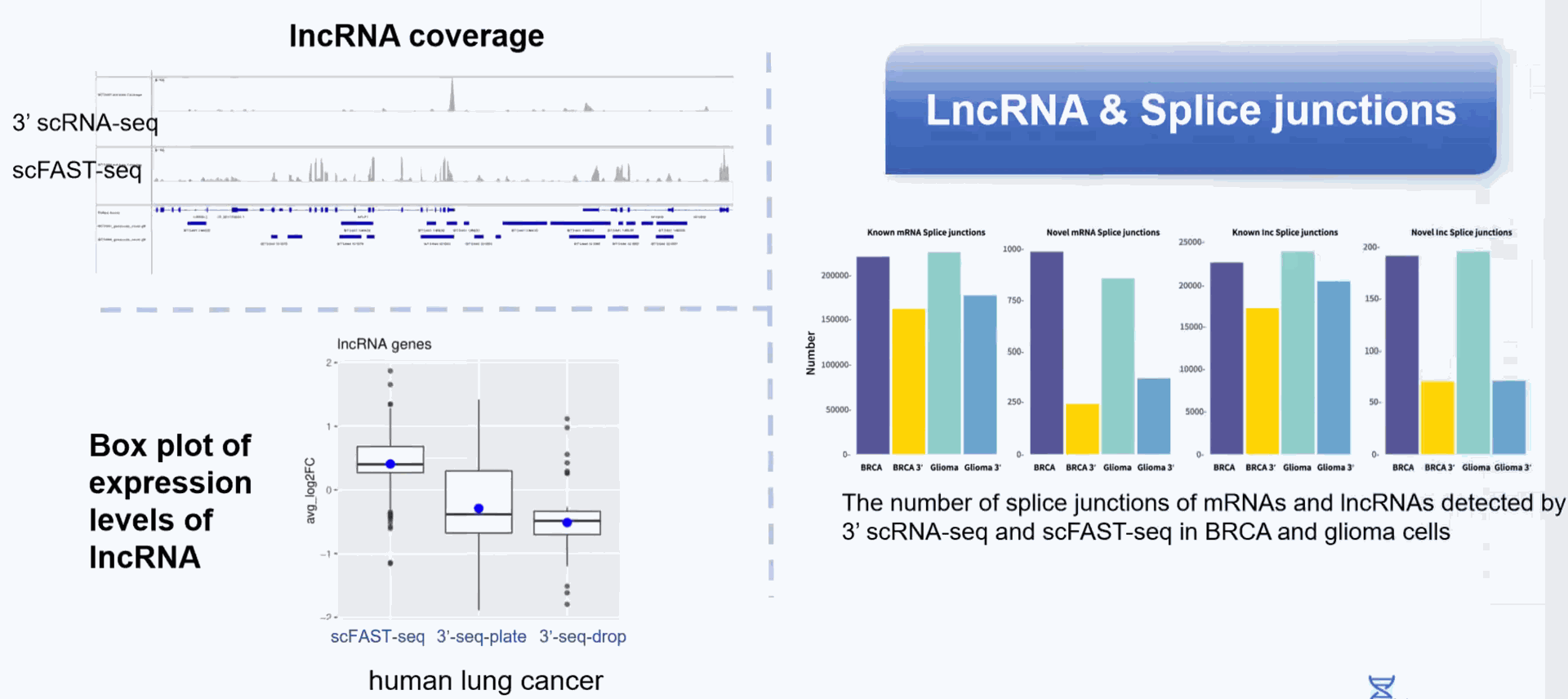
Better Capturing non-coding RNA: In addition to mRNA, non-coding RNA without 3' polyA tails can also be captured.
Crossing the species barrier: Enable the detection of viruses and prokaryotes.
Improving analysis accuracy: Allow the detection of isoform switching events of mRNA and IncRNA.
Applications
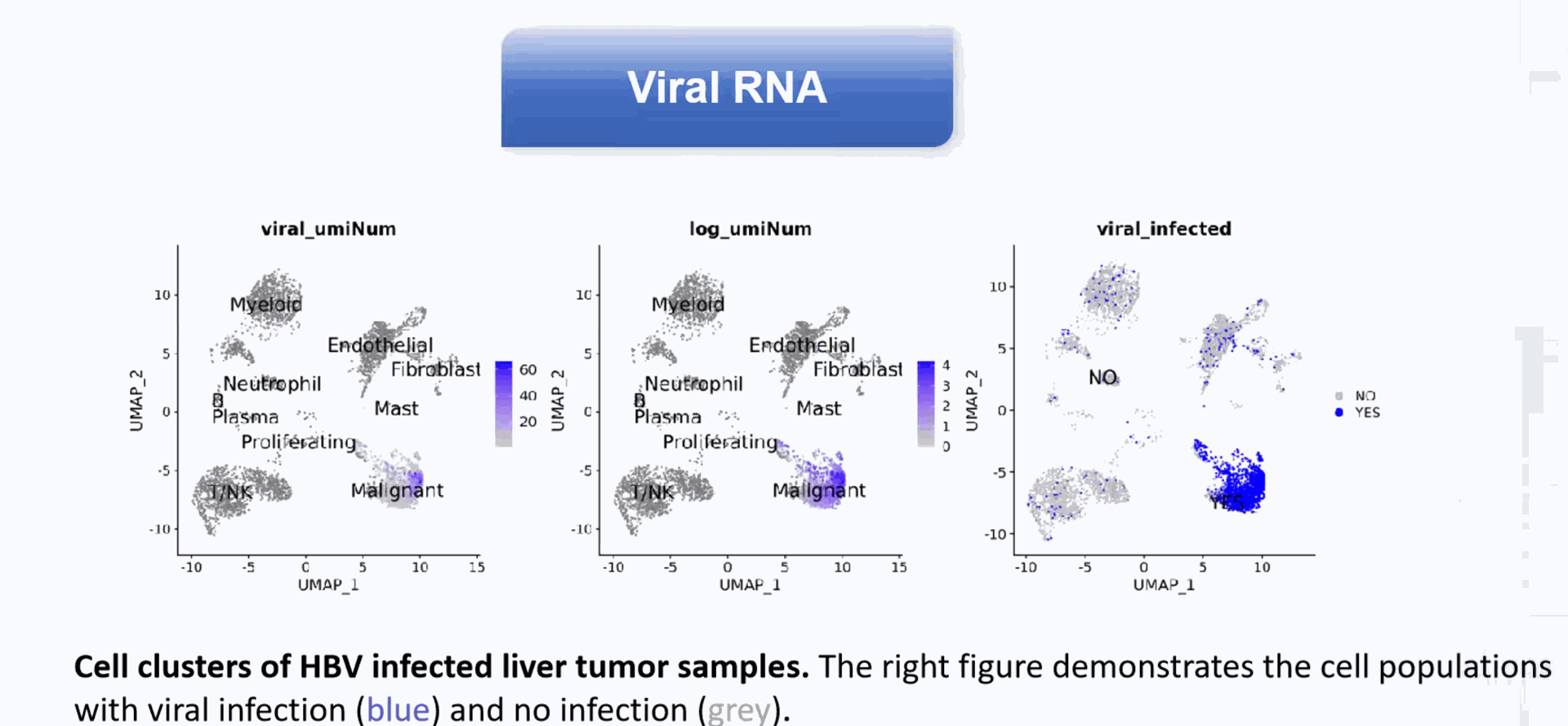
Detection of more splice junctions
Identification of non-polyA tailed viral infected cell clusters
Flexibility to incorporate a panel for detecting target genes with high
Achieving effective rRNA depletion: the inclusion of an rRNA blocker enables efficient depletion of rRNA, delivering higher proportion of effective data.
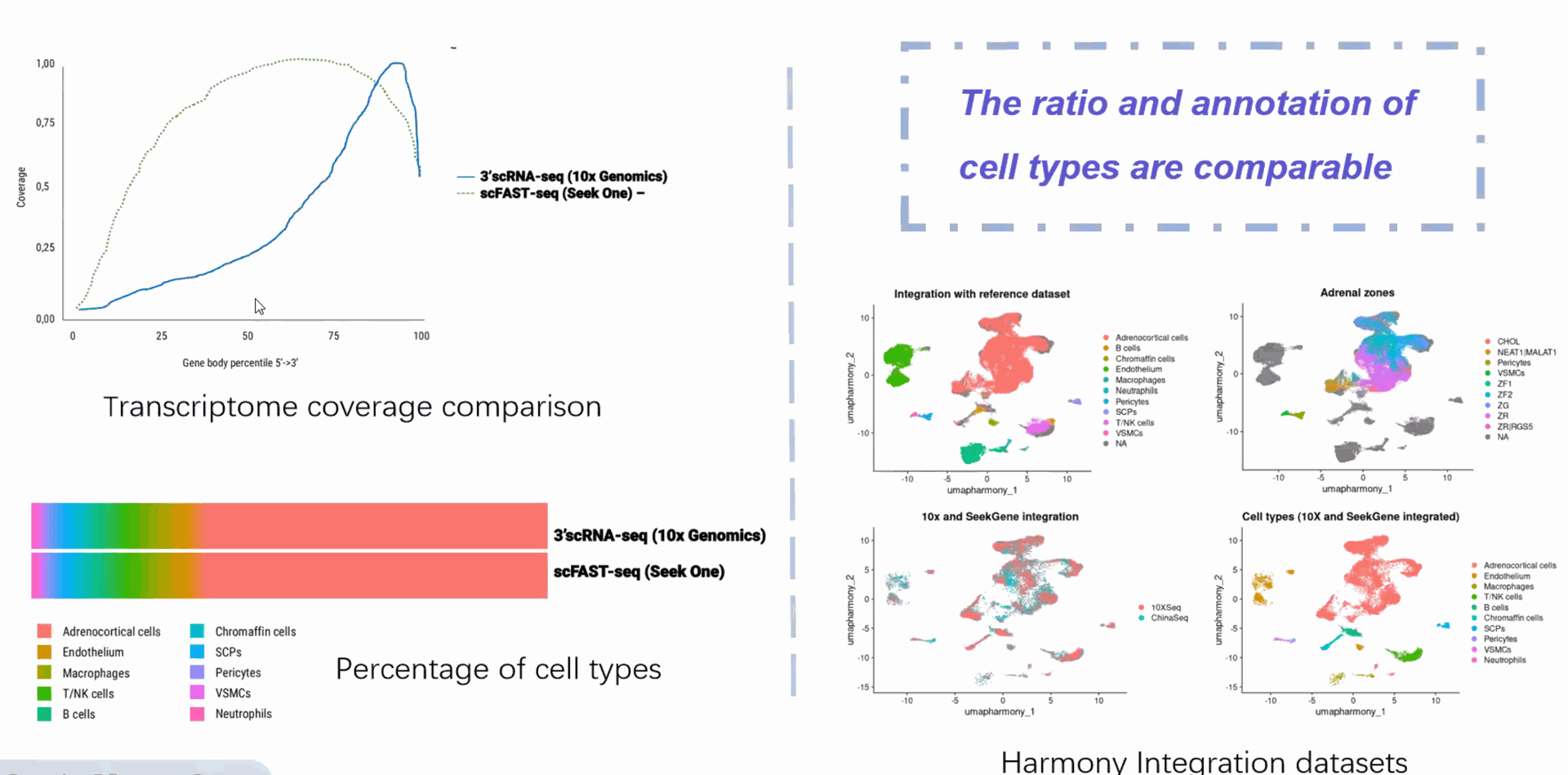
Compared to conventional 3' scRNA-seq, scFAST- seq has distinct advantages in detecting non- polyadenylated transcripts, transcript coverage length, and identification of more splice junctions. With target region enrichment, scFAST-seq can simultaneously detect somatic mutations and cell states in individual tumour cells, providing valuable information for precision medicine.
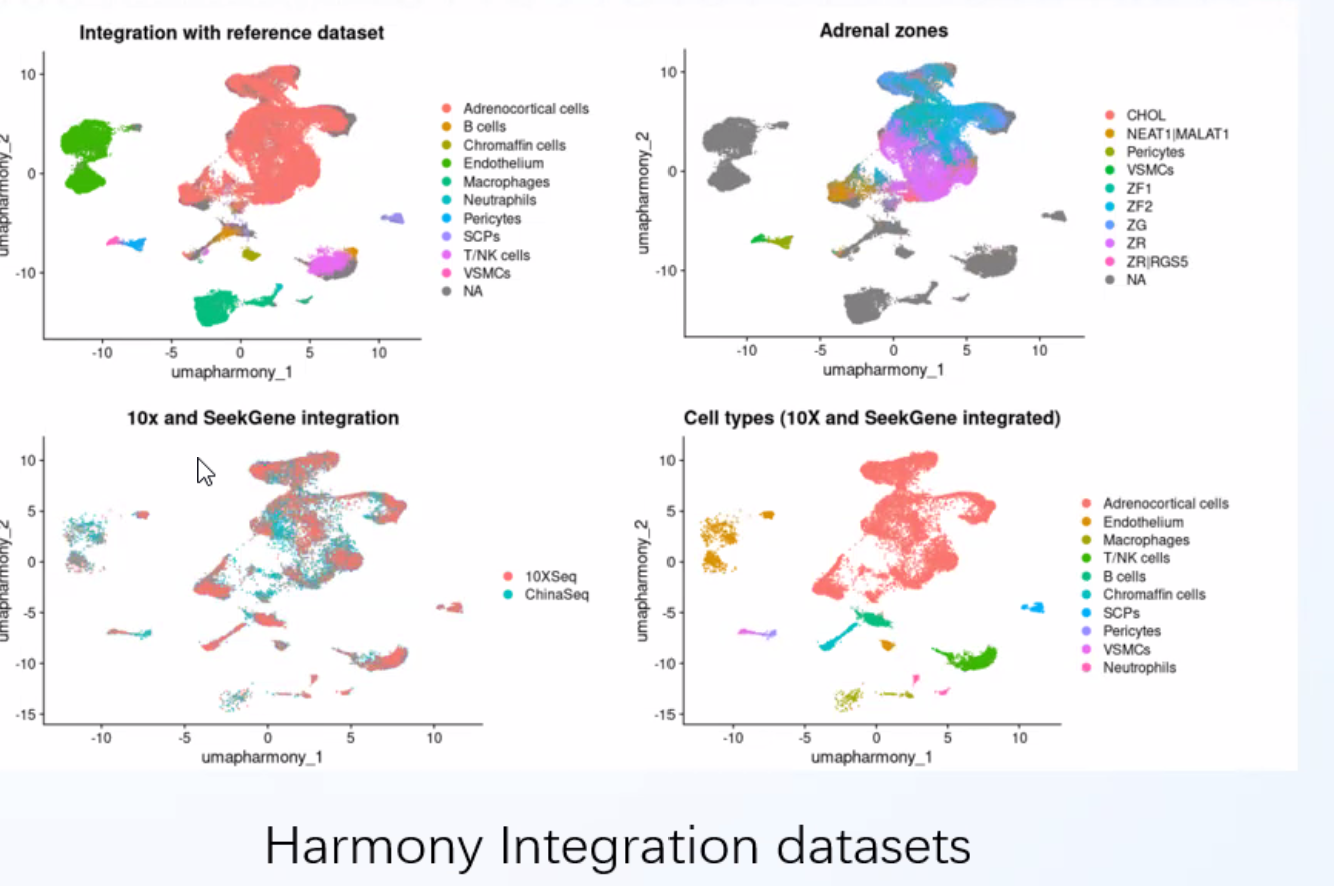
Clearer and more accurate cell trajectory inference compared to the 3' platform
scFAST-seq data can be integrated with 3' data
Product | Cat.No. | Specification |
| SeekOne® DD Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) | K00801-08 | 8 tests |
PRODUCTS
Single Cell Immune Profiling Kit
—— Reveal the immune specificity with complete V(D)J sequences
SeekOneDD single-cell immune profiling (5' transcriptome & immune repertoire sequencing), proposed by Beijing SeekGene BioSciences Co., Ltd., achieves single-cell partitioning and labelling by the microfluidic system and barcoded beads. The library of both full-length TCR/BCR sequences and mRNA sequences can be constructed from a single sample, followed by high-throughput sequencing and bioinformatics analysis.
The SeekOne DDSingle Cell Immune Profiling Kits include the SeekOne
DD Single Cell 5’ Transcriptome-seq Kit and the SeekOne
DD TCR/BCR amplification Kit. With this kits, the full-length sequence of V(D)J, the pairing information of α chain (heavy chain) and β chain (light chain) of TCR(BCR) and the transcriptome information can be obtained simultaneously. It can be used to investigate T/B cell heterogeneity, TCR/BCR diversity, antigen specificity, vaccine development, immunetherapy screening, etc.
1.Single cell capture and labelling
SeekOneDD single cell immune profiling products are based on the principle of microfluidic technology. Cells and barcoded beads are added separately and then react in the carrier oil to form emulsion droplets in the -
shaped channel. After capturing mRNA tagged with barcoded beads, two separate libraries (gene expression & immune repertoire) can be constructed from a single sample.
Core technology and workflow
Compatible Instrument
SeekOneDD
SeekOneDigital Droplet System
Single Cell 3'Transcriptome-seq kit
SeekOneDigital Droplet System
Reagent
Instrument
Software
SeekOneDD
Single Cell Immune Profiling Kit
SeekOneDD
Single Cell Full-length RNA Sequence Transcriptome-seq kit
Up to 90%of reads mapped to any V(D)J gene
Rapid generation of 150000 water-in-oil droplets in 3 minutes
Flexible running of 1~8 samples in parallel
High cell capture rates of up to 65%
Efficiently capture 500-12,000 cells per channel
Low doublet rates of under 0.3% per 1,000 cells
2.Library construction
Product Features
Full length
Complete coverage of the full-length V(D)J sequences. Analyze all V(D)J sequences, including CDR3 sequences, for more comprehensive information
True alignment
True representation of α/β (light/heavy) chain pairing information of TCR (BCR). Reports pairing information from the same cell to reveal receptor-antigen specificity of the immune cells
Efficient capture
Efficiently capture V(D)J sequences with an alignment rate greater than 90%. Obtain more accurate and authentic capture data
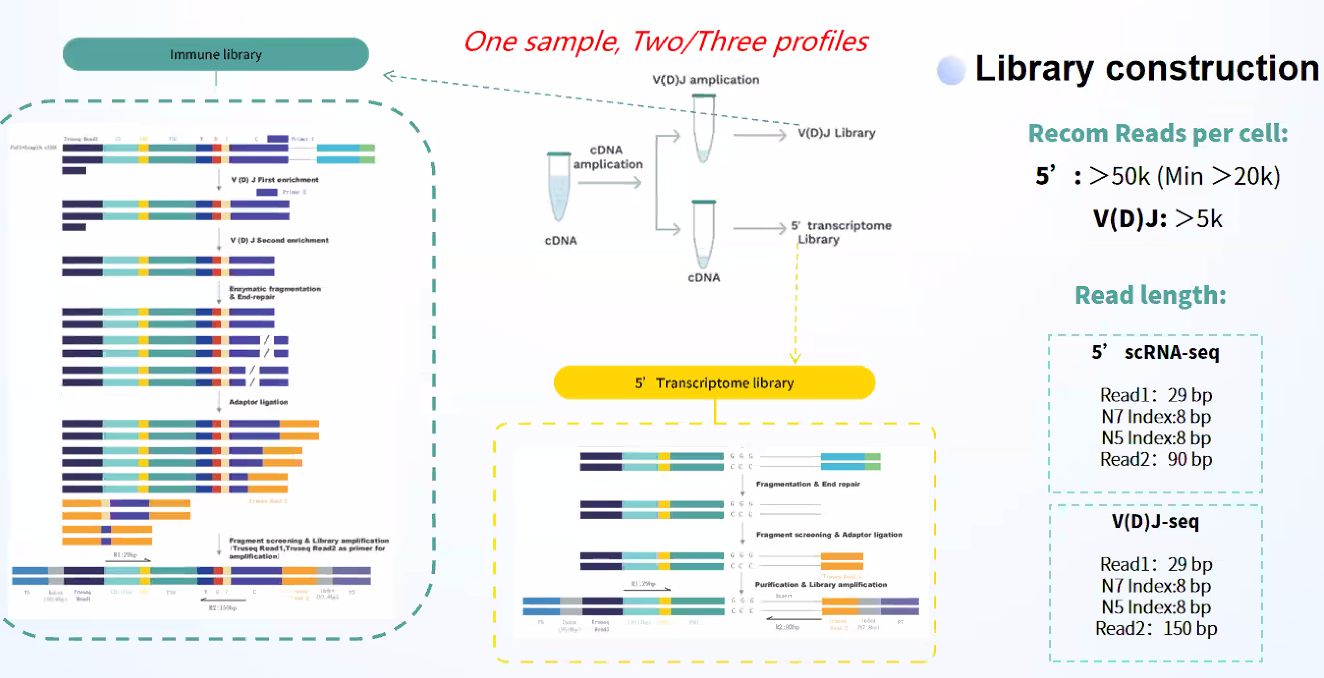
One sample with two profiles
Simultaneous profiling of immune repertoire (BCR/TCR) and 5’ gene expression Multiple libraries are constructed from a single sample, which is more economical and suitable for samples with limited amounts, such as needle biopsy samples.
Order Information
Product Name | Cat.No. | Specification |
SeekOne® DD Single Cell 5’ Transcriptome-seq Kit | K00501-08 | 8 tests |
SeekOne® DD Single Cell TCR Enrichment Kit (Human) | K00601-08 | 8 tests |
SeekOne® DD Single Cell BCR Enrichment Kit (Human) | K00701-08 | 8 tests |
SeekOne® DD Single Cell TCR Enrichment Kit (Mouse) | K01101-08 | 8 tests |
SeekOne® DD Single Cell BCR Enrichment Kit (Mouse) | K01201-08 | 8 tests |
SeekOneDigital
Droplet System
Fast
Rapidly generates 150,000 water-in-oil droplets in 3 minutes, reducing cell loss.
Efficient
Up to 12,000 cells can be captured in a single channel. Flexible running of 1~8 samples in parallel.
Accurate
Low doublet rates of less than 0.3% per 1,000 cells; equipped with a temperature control system to eliminate the effects of environmental temperature differences.
Compatible
Highly compatible system with a real-time remote upgrade can be used with comprehensive SeekOneproduct lines to meet a variety of scientific research needs.
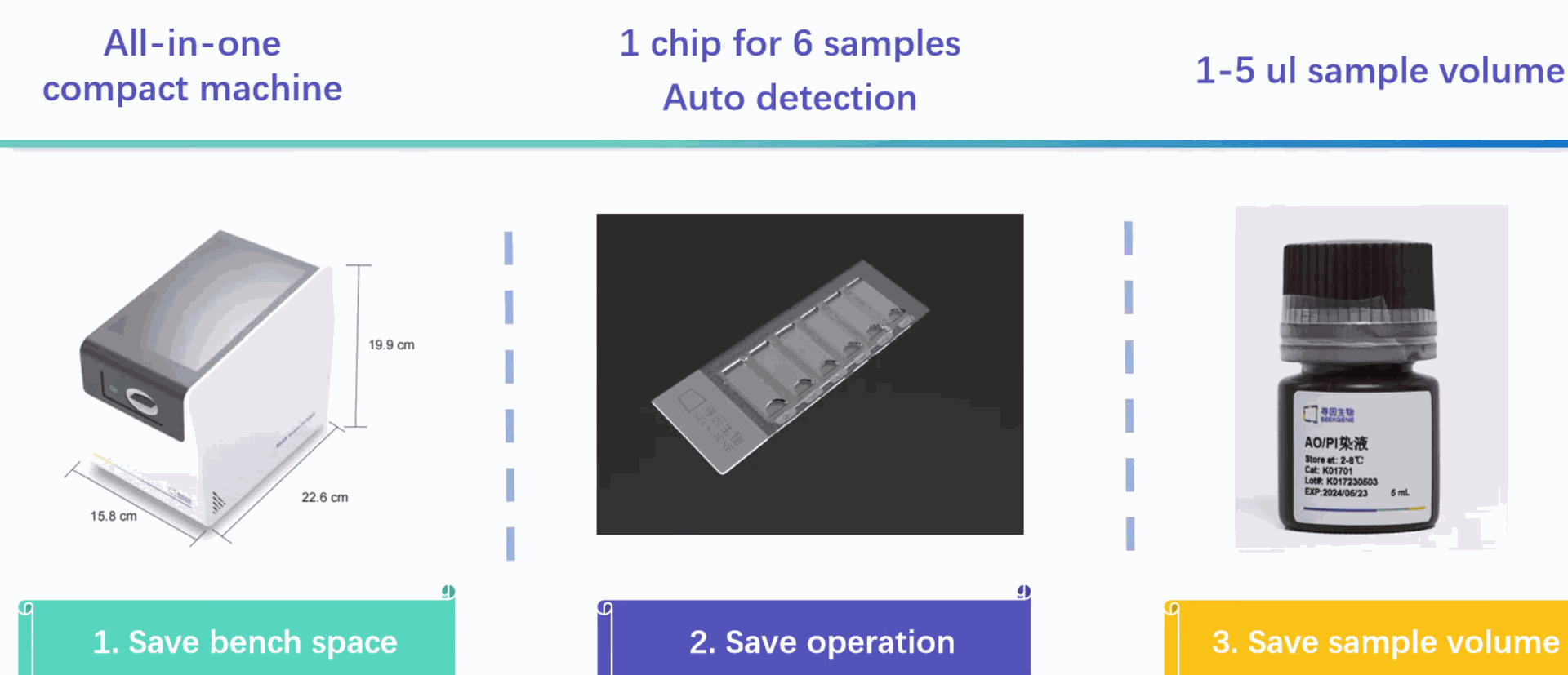
SeekOneDD Chip Holder
Chip Holder, used with SeekOne® digital droplet
system and supporting chips
Product Features
Placed Chip
Placed Chip (Chip P for short), placed in a Chip Holder (8 Chip Ps are attached to each instrument);
When the sample throughput is less than 8, Chip P is placed in the wells
User-friendly
Small instrument with large screen; 7" + 45° view touch interface makes “one-click” operation more convenient and comfortable.
SeekOne Digital Droplet System
SeekSoul Tools
Reagent
Instrument
Software
SeekOneDD
Single Cell 3 'Transcriptome-seq kit
SeekOneDD
Single Cell Immune Profiling Kit
SeekOneDD
Single Cell Full-length RNA Sequence Transcriptome-seq kit
Cells and barcoded beads are added separately and then react in the carrier oil to form emulsion droplets in the -shaped channel. After that, mRNA molecules released by the cells are captured by oligo(dT) on the barcoded beads.
Workflow
The process begins with the collection of single cell suspension. After single cell partitioning, capture and labelling, a single cell library compatible with Illumina and MGI sequencers is constructed for high-throughput sequencing. Data can be processed using SeekSoul® Tools ——the efficient data analysis software to explore cell heterogeneity.
Product Specifications
Flexible cell diameter of 5~40 μm
Rapid generation of 150000 water-in-oil droplets in 3 minutes
Efficiently capture 500-12,000 cells per channel
Flexible running of 1~8 samples in parallel
High cell capture rates of up to 65%
Low doublet rates of under 0.3% per 1,000 cells
Cost-saving
Vacant channels can be occupied by Place Chip without wasting chips.
Instrument | Cat.No. |
| SeekOne® Digital Droplet System | M001A |
SeekOne Chip Holder
SeekOne Chip Holder is the chip holder of the SeekOne
MM Single Cell Transcriptome Kit and
is used in conjunction with the SeekOne Chip.

Chip fixture
Product Performance
1 to 4 chips can be placed at a time, and a single operator can process 1 to 4 samples simultaneously.

SeekMate Tinitan
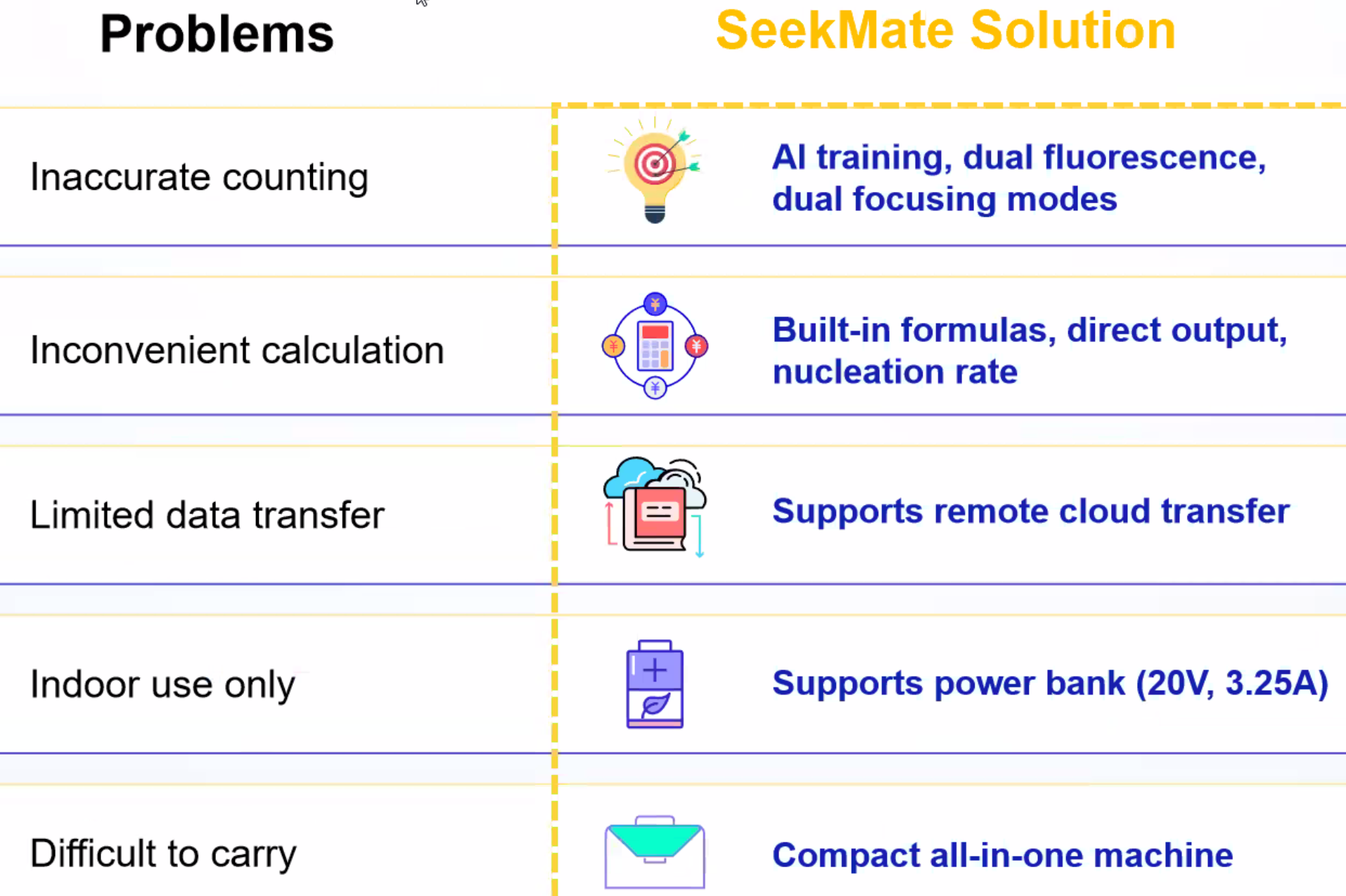
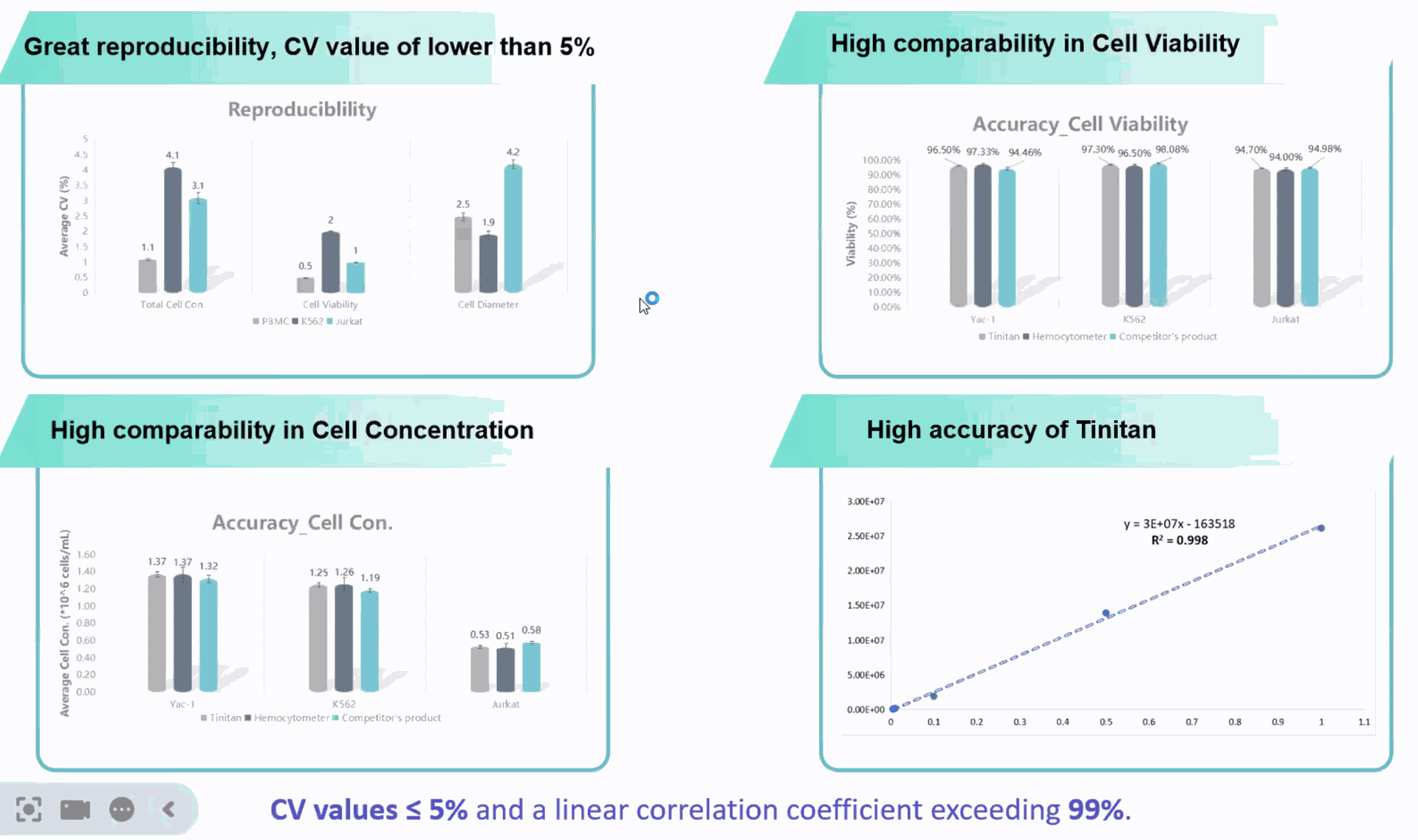
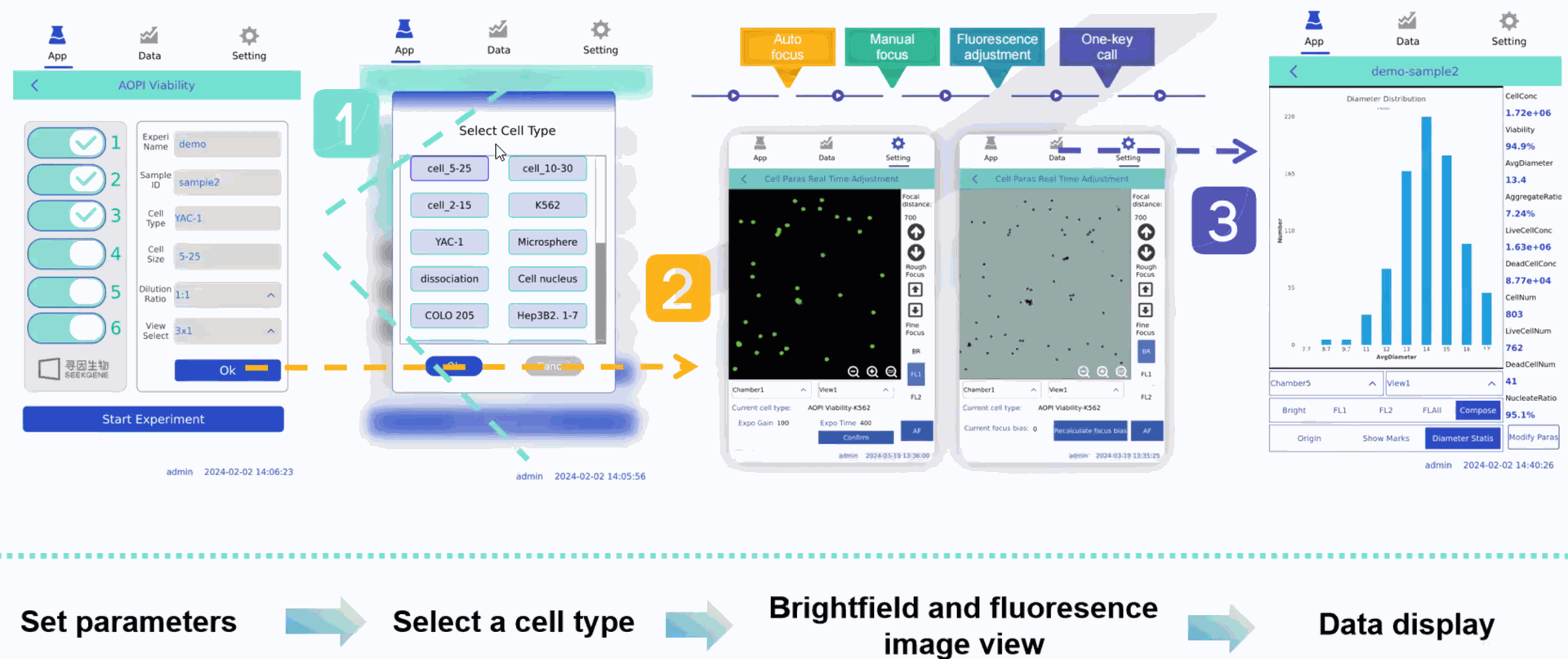
SeekMate Tinitan Fluorescence Cell Analyzer (SeekMate Tinitan FL) is an efficient fluorescence cell analyzer independently developed by SeekMate Biotechnology based on bright field and fluorescence field dual modes. This product uses the latest optical components with dual focusing modes to obtain clear pictures, and introduces AI deep learning. Personalized parameter adjustment provides accurate and stable cell counting solutions for more cell types (including primary cells and cell nuclei). At the same time, the machine is lightweight and compatible with mobile power supplies, which is convenient to carry out and can be used to perform cell counting in any cell laboratory. The device is compact and fully functional. It is a trustworthy handheld "counting expert" with accurate counting, information dimension expansion, and flexibility and convenience.

Fluorescence cell analyzer

Product Advantages
Reagent test kit
equipment
software

- SeekMate Dissociation Kit
- SeekOne
DD
- Single Cell 3' Transcriptome Kit
- SeekOne DD
- Single Cell Whole-Sequence Transcriptome Kit
- SeekOne
DD
- Single Cell Immunoassay Kit
- SeekOne
DD
- FFPE Sample Single Cell Transcriptome Kit
- SeekOne
Chip Holder
- SeekOne
Digital Droplet Meter
- SeekSoul CMDB Database
- SeekSoul Tools Analysis Software
- SeekOne
MM
SeekMate Tinitan Fluorescence Cell Analyzer
SeekSoul Tools
Products & Technologies
Analysis software
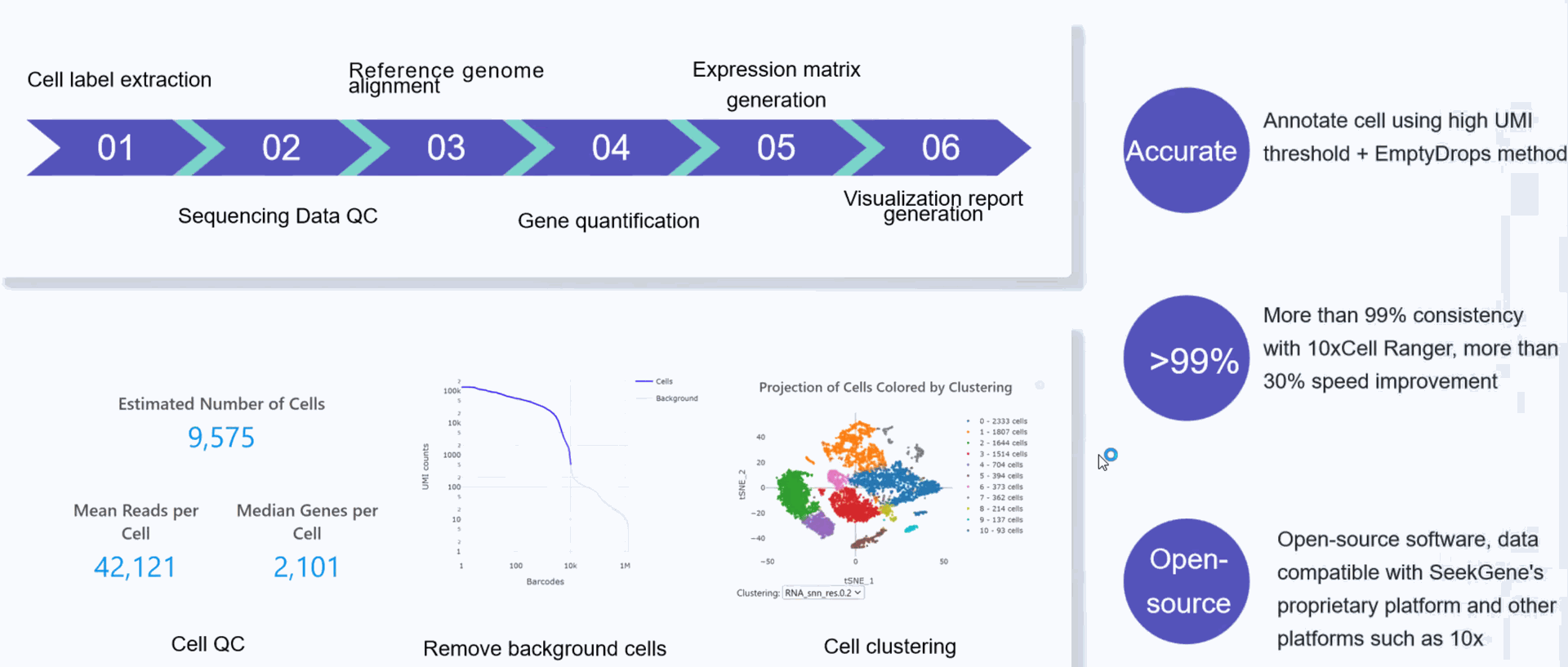
SeekSoul Tools is a set of software independently developed by SeekSoul Biotechnology to process single-cell transcriptome data. It is used to identify cell label barcodes, compare and quantify, obtain cell expression matrices that can be used for downstream analysis, and then perform cell clustering and differential analysis. The product not only supports the output data of the SeekOne series of kits, but also supports various custom design structures through the description of barcodes.
Analysis Process
Is SeekSoul Tools compatible with third-party software such as Seurat?
Frequently asked questions
SeekSoul Tools is a single-cell data analysis software independently developed by SeekSoul. It is used in conjunction with the SeekOne®MM/DD single-cell transcriptome library kit. It can only be used after installation under the Linux system. The customer needs to have computing resources and a certain bioinformatics foundation. This software package can complete the basic quality control of offline data. The software has good performance and is compatible with data from a variety of single-cell technology platforms. In addition, the cell-gene expression matrix obtained by SeekSoul Tools analysis can also be directly imported into third-party software such as the Seurat software package for subsequent analysis.
SeekSoul CMDB
SeekSoul CMDB is a single-cell omics marker gene database established by SeekSoul Biotechnology. It contains the marker genes used to annotate cell types in currently published single-cell sequencing articles on diseases and healthy tissues. Combined with the automatic annotation software developed by SeekSoul Biotechnology, it can assist in cell type identification and annotation.
database