SeekGene SeekOne
- ® DD Single Cell Full-lengthRNASequence Transcriptome-seq (scFAST-seq) Product Overview
- SeekOne
- ® Digital Droplet (SeekOne
- ® DD) High throughput Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit, self- developed.
Seekone microfluidic control system
SeekOne is a powerful commercial tool for high-throughput whole transcriptome profiling. The scFAST-seq method makes use of innovative techniques
including semi-random primers, efficient reverse transcription, template swapping, and effective rRNA removal to build full-length RNA libraries of up to 12,000 cells. Compared to conventional 3' scRNA-seq, scFAST- seq has distinct advantages in detecting non- polyadenylated transcripts, transcript coverage
length, and identification of more splice junctions. With target region enrichment, scFAST-seq can
simultaneously detect somatic mutations and cell states in individual tumour cells, providing valuable
information for precision medicine. The kit is designed to be used in conjunction with
our SeekOne
® Digital Droplet System (SeekOne
® DD)
to complete the entire process from single-cell nucleic acid labelling to transcriptome library construction. When equipped with "SeekSoul Tools", our single-cell data analysis software, we provide a one-stop-shop single-cell transcriptome solution.
Highlights
Bring a powerful tool for studying tumour heterogeneity by mutation + gene expression-profiling at the single cell level. Study isoforms generated by alternative splicing and long non-coding RNA in gene structure. Explore the pathogenic mechanisms associated with the expression characteristicsof cell types mapped by mutant genes. Detect polyA-tailed and non-polyA-tailedviral
RNA transcripts to analyze the specificity of virus-infected cell populations, virus-host cell
interactions, and microenvironmental changesinduced by viral infection. Facilitate basic R&D, drug development, non-clinical and clinical evaluation, manufacturingquality control, etc. Break the single-omics limitation of providingonly one dimension of single cell information.
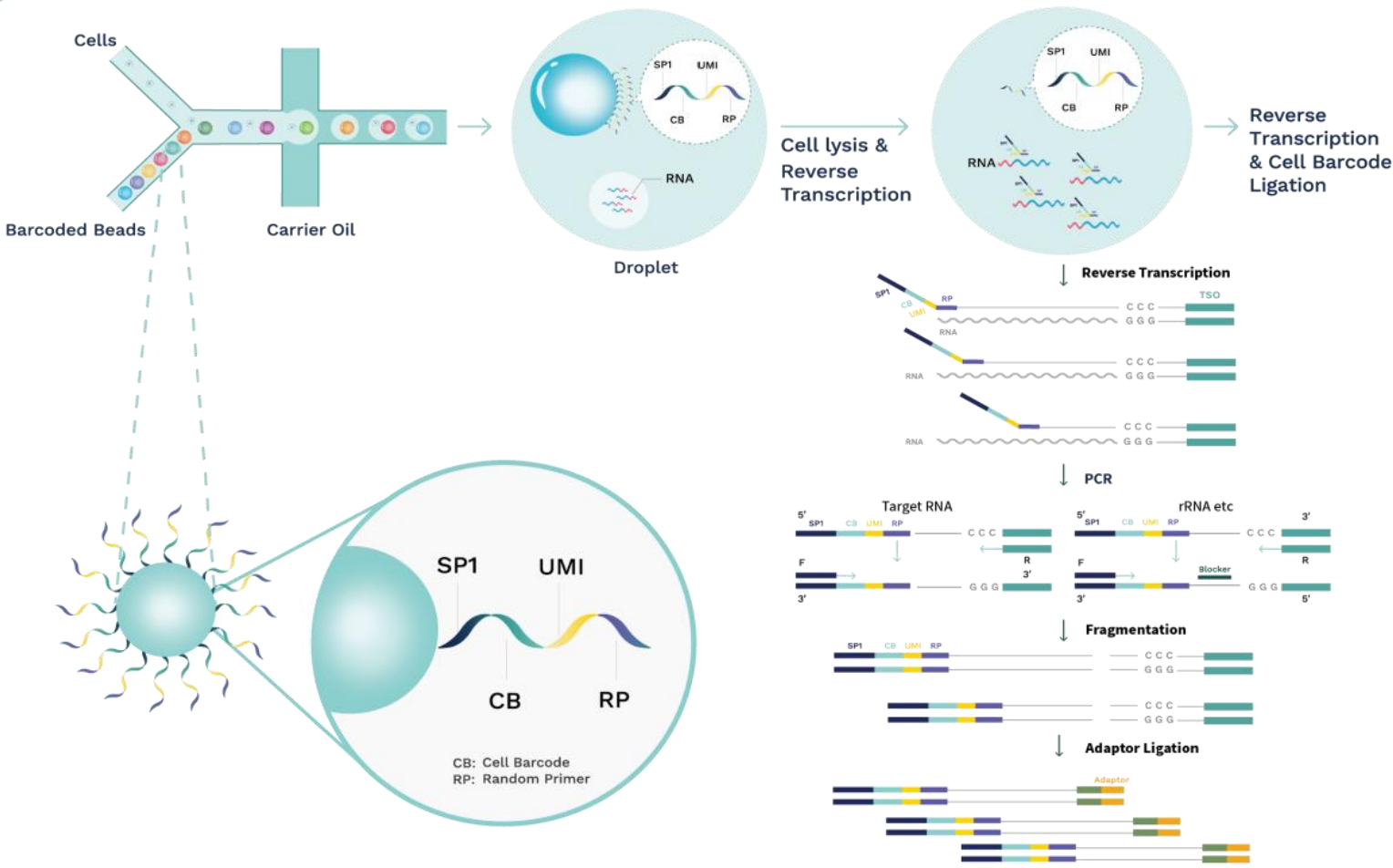
Figure 1. The core technology for Single Cell Full-length mRNA Sequence Transcriptome Sequencing (scFAST-seq).
2 Core Technology
Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) allows full-length
transcriptome analysis of thousands of fresh or
fixed cells in a single experiment. In contrast to the
current single-cell transcriptome assays based on
oligo-dT reverse transcription, the method used in
scFAST-seq provides coverage of the whole
transcriptome. The sensitivity of the scFAST-seq
technique assays depends primarily on the
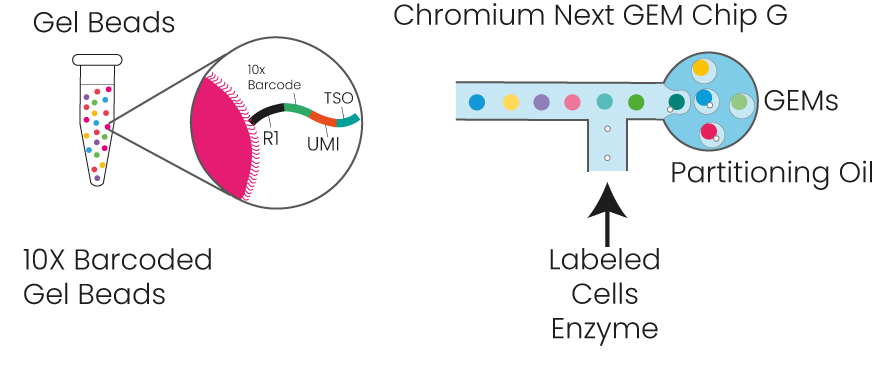
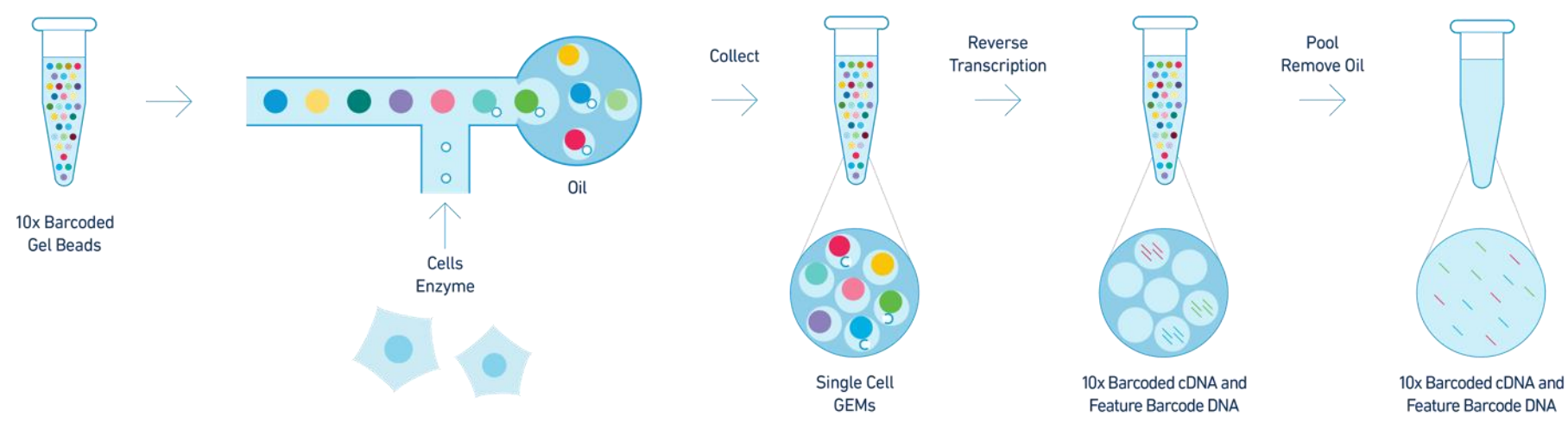
efficiency of reverse transcription, cDNA enrichment, and ribosome depletion. First, cells and barcoded beads are added and react
to generate emulsion droplets in the -shaped
channel of the SeekOne
® Digital Droplet System
(Figure 1). Reverse transcription is then performed
by encapsulating the microbeads coupled to cell
labels and semi-random primers with individual cells in a single droplet, resulting in cell-labelledcDNA fragments. Once the reverse transcriptionof each cDNA is complete, a template transitionsequence (template switch oligo, TSO) is addedtothe 3' end of the cDNA using the end-transferaseactivity of the reverse transcriptase. The barcodedcDNA is then amplified in vitro, where the ribosomal
RNA (rRNA) is depleted. Finally, a portion of thecDNA is fragmented, end-repaired, and ligatedtoasequencing adaptor. The library is constructedandamplified using the unique dual-indexed polymerasechain reaction (PCR) primers for sequencing onIllumina or MGI platforms. In this way, the cDNAamplified by PCR would reflect both polyA andnon-polyA RNA sequences.
Product Features
Genotype to
phenotype
Simultaneously obtain single-cell mutation and corresponding gene
expression information, establishing the relationship between
genotype and phenotype
- Accurate single-cell
- transcriptome
- Achieve full-length transcriptome detection, without being limited by
- the 3’ or 5’ end.
- Crossing the species
- barrier
- Enables the detection of viruses and prokaryotes. Breaking the species
- limits
- In addition to mRNA, non-coding RNA without 3’ polyA tails can also
- be captured.
- Improving analysis
- accuracy
- Allows the detection of isoform switching events of mRNA and
- lncRNA
Product Specifications
- Currently applicable species: human and mouse Fraction of Reads Mapped to Middle Genebody is greater than 55%
- Fraction over 0.2 mean coverage depth of ACTB gene is higher than 70%
- Rapid generation of 150,000 water-in-oil droplets in 3 minutes Efficiently capturing 500-12,000 cells per channel
- Flexible running of 1~8 samples in parallel
- Cell size flexibility: cell diameter of 5~40 μm
- High cell capture rates of up to 65%
- Low doublet rates of under 0.3% per 1,000 cells
3 Workflow Steps
Tissue sample dissociation Single cell partition
and labeling
Full-length RNA
sequence library construction High-throughput sequencing Data analysis
Figure 2. SeekOne
® DD Single Cell Full-length mRNA Sequence Transcriptome Sequencing (scFAST-seq) Workflow. SeekOne
® DD scFAST-seq
overcomes the restriction of 3' or 5' scRNA-seq that only sequences at the ends of mRNA can be amplified. By using microfluidic digital droplets and barcoded beads coupled with semi-random primers, the scFAST-seq kit can achieve random capture of the full-length RNA sequence
transcriptome, allowing the detection of both coding and non-coding RNA. ScFAST-seq data can be processed with SeekSoul® Tools ——our efficient data analysis software for comprehensive information on mutations, fusions, and alternative splicing. Data Presentation
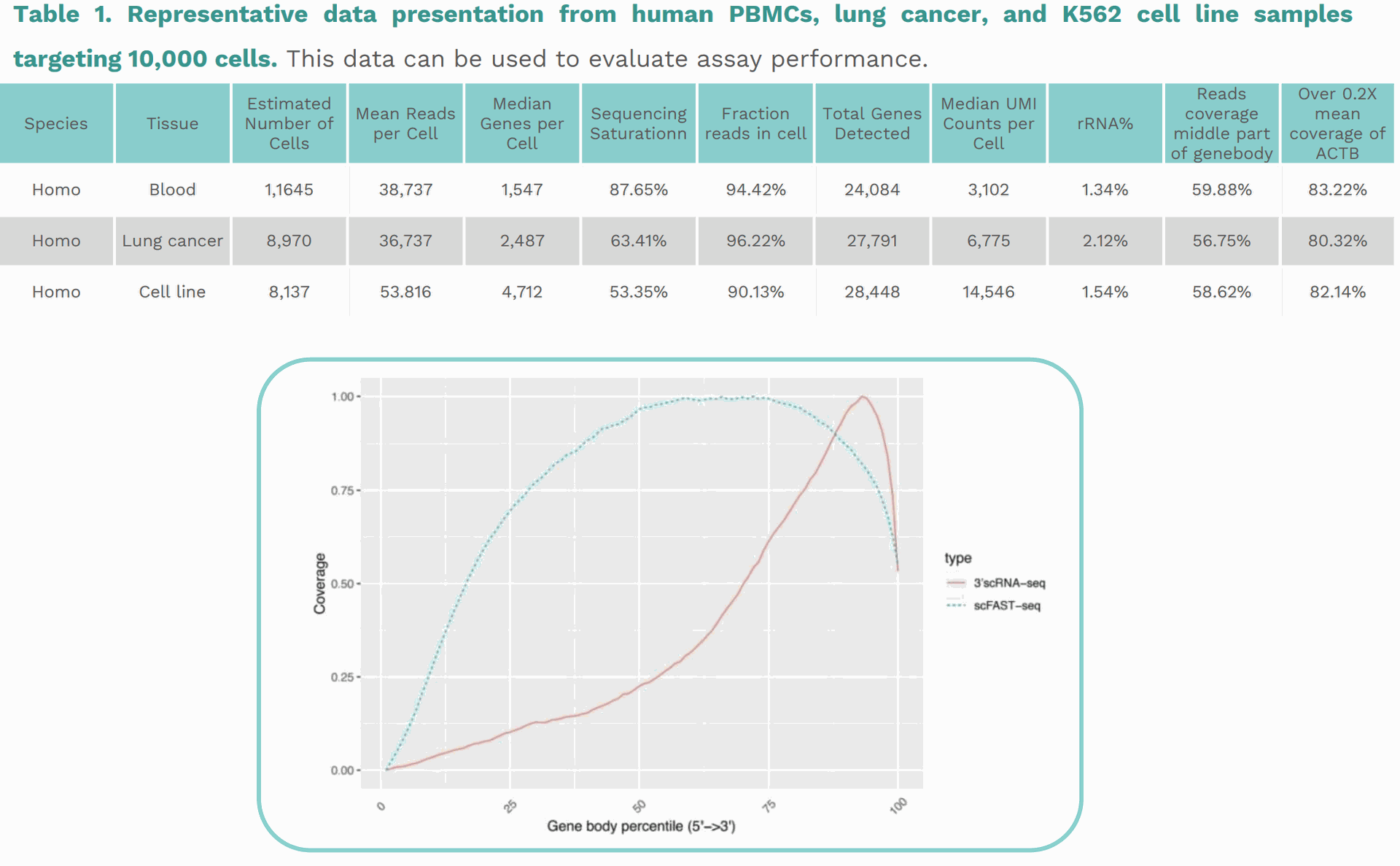
Table 1. Representative data presentation from human PBMCs, lung cancer, and K562 cell line samplestargeting 10,000 cells. This data can be used to evaluate assay performance. Species Tissue Estimated Number of Cells Mean Reads per Cell Median Genes per Cell Sequencing Saturationn
Fraction
reads in cell Total Genes Detected Median UMI Counts per Cell rRNA%
Reads coverage middle part of genebody Over 0.2Xmeancoverageof ACTBHomo Blood 1,1645 38,737 1,547 87.65% 94.42% 24,084 3,102 1.34% 59.88% 83.22%Homo Lung cancer 8,970 36,737 2,487 63.41% 96.22% 27,791 6,775 2.12% 56.75% 80.32%Homo Cell line 8,137 53.816 4,712 53.35% 90.13% 28,448 14,546 1.54% 58.62% 82.14%Figure 3. Comparison of transcriptome coverage between 3’ scRNA-seq and scFAST-seq. The coverage of 3’ scRNA-seq is significantly skewedtothe left of the 3' end. In contrast, the scFAST-seq is evenly distributed across the gene body percentile, indicating a full-length coverage of theentire gene body and the capture of unspliced and splice junction regions with less bias.
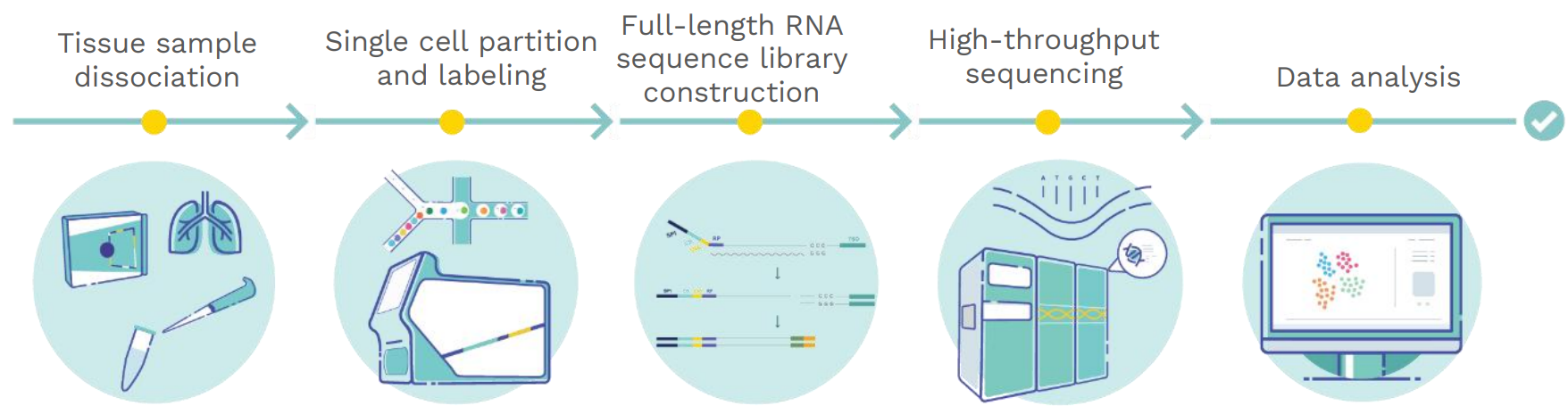
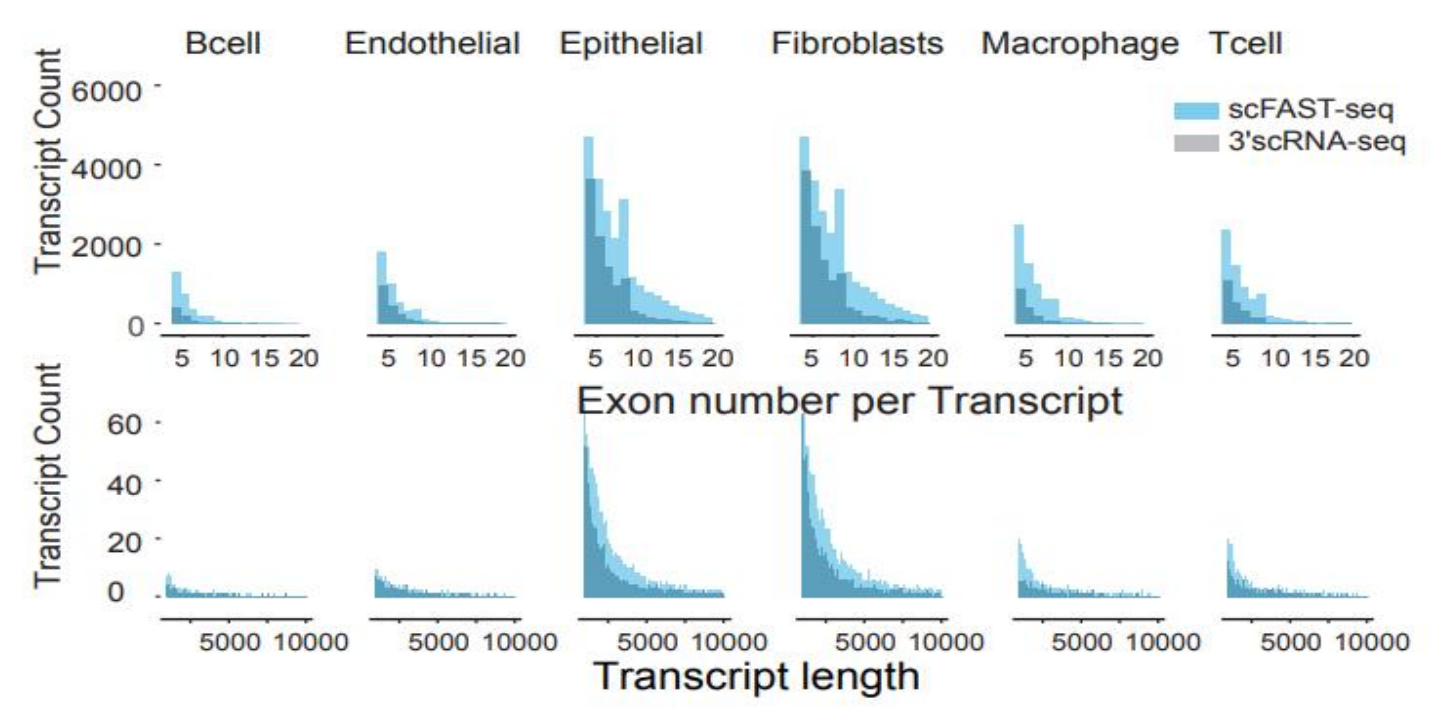
Figure 4. The number of transcripts of each cell in different cancer and PBMC samples. Blue represents the full-length technique scFAST-seq andgrey denotes the 3 ' scRNA-seq. scFAST-seq detected significantly more transcripts than 3’ scRNA-seq in glioma and breast cancer tissue. A.
B.
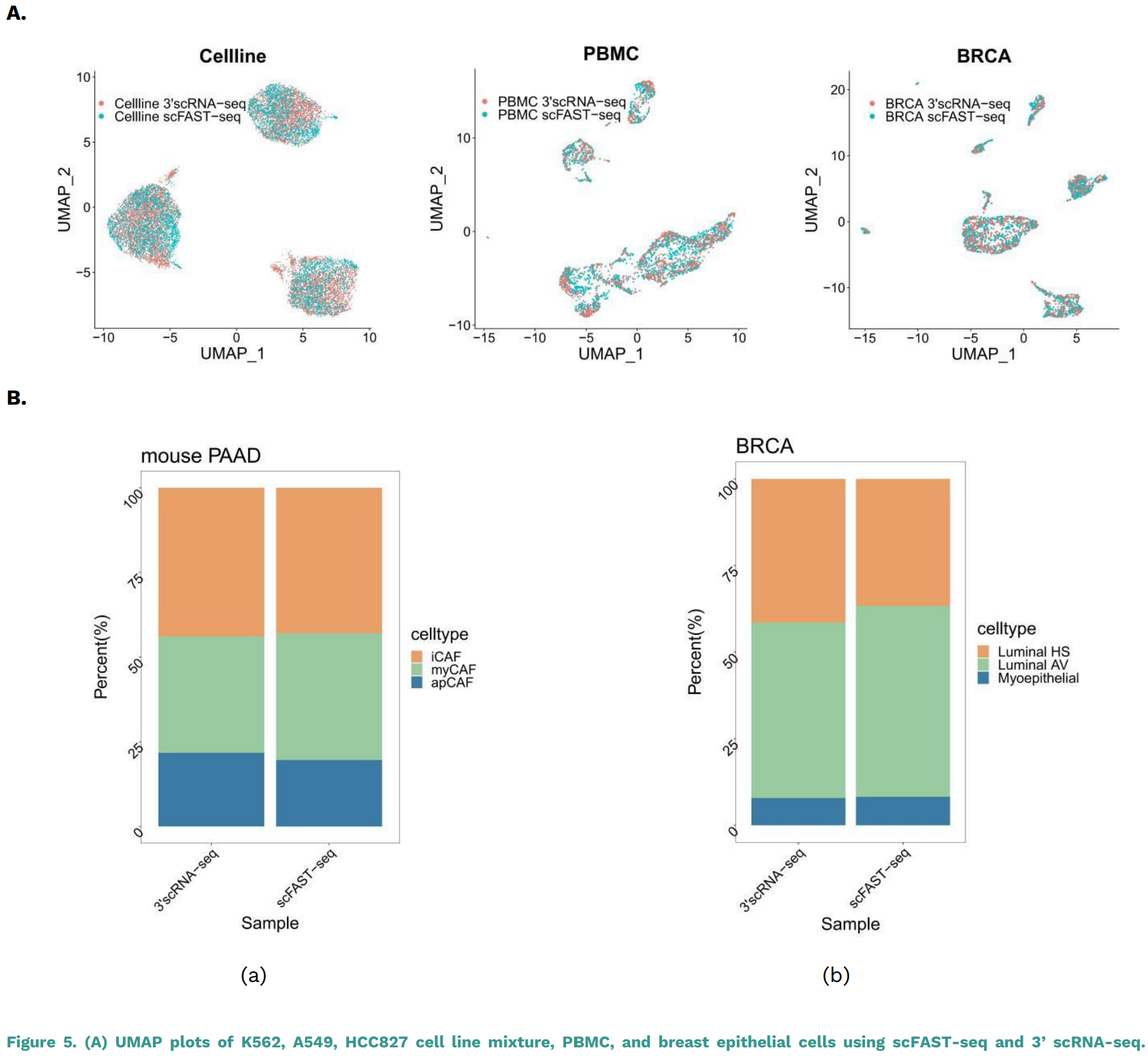
(a) (b) Figure 5. (A) UMAP plots of K562, A549, HCC827 cell line mixture, PBMC, and breast epithelial cells using scFAST-seq and 3’ scRNA-seq. By applying the typical correlation analysis method in the Seurat package to integrate and visualize the scFAST-seq and 3’ scRNA-seq data, it was
found that the clustering and relative positions of cells in the two types of data in the three samples were highly overlapping in the two- dimensional UMAP. (B) Cell subtype proportions in mouse fibroblasts (a) and epithelial (b) detected by 3’ scRNA-seq and scFAST-seq. The subtypeproportions of fibroblasts in pancreatic cancer and epithelial cells in breast cancer indicate a high consistency in cell proportions betweenscFAST-seq and 3’ scRNA-seq data, such as Luminal HS, Luminal AV, and Myoepithelial.
5 Applications
The single-cell FAST-seq allows access to more dimensions of information: Mutation
SeekOne
® scFAST-seq provides two dimensions
(mutation & gene expression) of single cell
information, overcoming the restrictions of single- omics (Figure 6). The simultaneous two separate profiling of mutations + expression at the single cell
level creates a novel tool for a wide range of studies, including precise identification of tumour cells, clonal evolution, mutation mapping, and
therapeutic evolution. In terms of co-mutation, it
helps to evaluate the different tumour co-mutationstrategies and to investigate the mechanisms of co-mutation-driven molecular dependencies. Furthermore, it can be used to explore the
expression characteristics of cell types with mutant
genes or changes in signaling pathways. WithscFAST-seq, differences in mutation and geneexpression profiles between primary and metastatictumour sites can also be revealed.
Figure 6. Mutation detection in a hybrid sample of K562 (BCR-ABL1 fusion), HCC827 (EGFR 19del), and A549 (KRAS G12S) cell lines. Data fromthescFAST-seq and targeted enrichment libraries allowed clear differentiation of each cell type and accurate detection of the targeted mutant genes
in each cell line. A549: KRAS G12S (mRNA 5' end); HCC827: EGFR 19del (At the middle of the gene); K562: BCR-ABL1 fusion (At the middle of the gene)
Alternative splicing
Compared to 3' scRNA-seq, scFAST-seq yields a
significantly higher number of alternative splicing
sites for novel and known mRNA/lncRNA, which
expands our understanding of transcriptome and
proteome diversity, the complexity of gene
expression and the underlying mechanisms of RNA
regulation (Figure 7). As alternative splicing mediates diverse biological processes throughout
the life span of organisms, scFAST-seq analysis canbe applied to cellular differentiation and organismdevelopment.

6
Figure 7. A bar chart plot of the number of known and unknown splice junctions of recognized mRNAs and lncRNAs in BRCA and glioma cells. scFAST-seq achieves even coverage of the entire gene body, thus allowing unbiased capture of unspliced regions and splice junctions. Comparedto 3’ scRNA-seq, the scFAST-seq provides more alternative splicing detection of known or novel mRNAs and lncRNAs. Alternative splicing sites for novel and known mRNA/lncRNA: scFAST-seq > 3’ scRNA-seq. Non-polyA tailed RNA
The semi-random primers of scFAST-seq enable the
capture of viral RNA genes with polyA tails or non- polyA tails to identify non-polyA viral infected cell clusters (Figure 8). This detection overcomes the
species limitations and facilitates studies in viral
immunology research, including the identification of
virus-infected cell populations, virus-host cell
interactions, changes in the microenvironment
induced by virus infections, exploration of viral
therapeutic targets, and characterization of geneexpression at different infection or treatment stages.
Figure 8. Identification of non-polyA viral infected cell clusters. For viral gene detection, the semi-random primers used in scFAST-seq cancapture viral RNA genes with polyA tails or non-polyA tails. The viral UMI in individual cells obtained by scFAST-seq can be linked back to the cell type annotation plot.
7 SeekOne Single Cell Full-length RNA SequenceTranscriptome-seq (scFAST-seq) Kit
The SeekOne DD Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit includes

- SeekOne®DD Chip S3 (Chip S3), Gasket, Carrier Oil, SeekOne
- ® DD scFAST-seq Barcoded Beads, amplification reagents, library construction reagents, and single-cell data analysis software (SeekSoul® Tools). SeekOne
- ® DD Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit
- Product Product code SeekOne
- ® DD Single Cell Full-length RNA Sequence Transcriptome-seq
- (scFAST-seq), 2 tests/8 tests K00801-02/K00801-08
- Product Components Component code SeekOne
- ® DD Chip S3 Kit, 2 tests/8 tests K00202-0201/K00202-0801
- SeekOne
- ® DD scFAST-seq Barcoded Beads Kit, 2 tests/8 tests K00801-0202/K00801-0802
- SeekOne
- ® DD scFAST-seq Reverse Transcription Kit, 2 tests/8 tests K00801-0203/K00801-0803
- SeekOne
- ® DD Library Construction Kit, 2 tests/8 tests K00202-0204/K00202-0804
- SeekOne
- ® DD scFAST-seq Cleanup Kit, 2 tests/8 tests K00202-0205/K00202-0805
Compatible Instrument
Compatible Instrument Product code SeekOne
® Digital Droplet System M001A
Compatible Software Compatible Software SeekSoul®Tools single-cell data analysis software FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES.
SeekOne
® DD scFAST-seq
-----Explore life with full-length sequence